Genes and QTLs for semolina and flour color
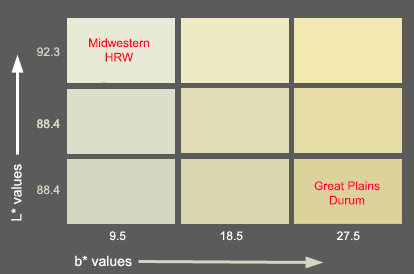
The colors of semolina and flours are expressed using the L* a* b* color system. L* is a measure of brightness, it can ranges from 0, completely non-reflective or black and 100, perfect white or total reflection. Bread wheat flours have reading values around 90, while semolina has lower values. The b* value is the blue-yellow chromaticity coordinate, it can go from -60, pure blue, to +60, pure yellow. Usual b* values for bread wheat flours are around 9.5. For semolina the higher the b* value the more yellowness. Good quality durums has a b* of aprox. 27.3 or more. The b* vlaue is the red-green coordinate
Cooked pasta has higher b* values, more than 41 for good quality durums. There is another quality parameter related to pasta color, the color score, which combines L* and b* in one index. A spaghetti sample with a color score of 9.0 or higher is good. The range of the color score is 1 - 12.
Other factors that affect pasta color are ash content and the lipoxygenase activity, which has a negative effect being responsible for browning, and industrial factors such as milling conditions, extraction rate and pasta processing.
Tetraploid durum wheat (Triticum turgidum L. var. durum) is the source of the semolina used for manufacturing pasta products. As with bread wheats, there are many quality requirements that the durum wheats must meet. One of these is the yellow color of the semolina, which is correlated with the color of pasta. Consumers have traditionally associated a yellow color with higher quality. Current regulations in many countries ask for pasta products with a yellow color that originates from the wheat and not from any other additive.
Previous works showed that the chemical substances responsible for the yellow pigment(s) in endosperm were carotenoids of the xanthophylls type, mainly lutein in free form and sterified. However, Hentschel et al. (1) applying more sophisticated separation techniques found that the chemical nature of the yellow pigment in semolina is quite complex: they found that the carotenoids fraction accounted for only 30-50 % of the yellow pigment weight, while the rest were unidentified compounds.
Elouafi et al. (2) identified a QTL with a large effect on yellow pigment in the distal region of the long arm of chromosome 7B in a cross between the cultivated durum variety Omrabi5 and T. dicoccoides. Microsatellite markers Xgwm344-7B explained 53% of the variation in yellow pigment in this cross. A QTL with smaller effect was found on the distal region of chromosome arm 7AL. Microsatellite marker Xgwm63e explained 13% of the variation in yellow pigment. Omrabi5 contributed the alleles for high yellow pigment. Hessler et al. (3) identified a minor QTL for yellow pigment on chromosome 5A in the cross Jennah Khetifa x Cham 1. RFLP marker Xbcd926 explained 10.4% of the variation, and Cham 1 contributed the allele for high yellow pigment.
QTLs for yellow pigment in hexaploid wheat
A high level of yellow pigment is desirable in durum wheat but is an undesirable trait in bread wheat. Several studies have identified QTLs for yellow pigment in crossed between bread wheat varieties. The main results are summarized below.
| Arm | Cross | Explained variation | Marker | Ref. |
| 7AL | Schomburgk x Yaralinka | 60 % | STS from AFLP Xwua26.4 | 4-5 |
| 7BL | CD87 x Katepwa | 10 % | Xpsr680 | 6 |
| 2D | CD87 x Katepwa | 12 % | Xwmc25a | 6 |
| 3A | CD87 x Katepwa | 17 % | XksuB8 | 6 |
| 5D | Cranbrook x Halberd | (LOD 4.2) | -- | 6 |
| 7A | Cranbrook x Halberd | (LOD 4.2) | Xfba349-Xpsr121 | 6 |
| 3B | Sunco x Tasman | 20 % | Xgwm285-Xcdo583 | 6 |
| 5B | Sunco x Tasman | 12 % | Xgwm499 | 6 |
| 7A | Sunco x Tasman | 27 % | distal Xwmc346 | 6 |
QTLs for yellow pigment in tetraploid wheat
Pozniak et al. (7) found in durum cultiva Kofa that the Phytoene synthase 1 (PSY-B1) gene to be linked to a 7BL QTL for high semolina color and suggested that it might be a good candidate gene for this QTL. The enzyme phytoene synthase (PSY) appears to catalyze a rate-controlling step in the synthesis of phytoene, one of the first molecules in the carotenoid biosynthetic pathway.
The Y, Lr19 and Sr25 genes on chromosome arm 7EL

The distal region of the chromosome arm 7EL from Lophopyrum ponticum carries a gene that increases yellow pigment in the endosperm, known as gene Y, the leaf rust resistance gene Lr19 and the stem rust resistance gene Sr25, which confers resistance to the African race Ug99. [link to the Sr25 page]
The 7EL segment was reduced by homoeologous recombination (8), and different common wheat and durum wheat lines were obtained that can be used for breeding.
Markers for Lr19, Sr25 and Y
PSY-B1 specific primers
Zhang and Dubcovsky (9) developed a set of markers for the Psy-B1 alleles of the cross UC113 x Kofa
PSY1_BF3 5'- GTGGAACTTGCATGCTATACA -3'
PSY1_BR2 5'- GAACCTCAGGTTCACATTCC -3'
The annealing temperature can be set between 56ºC to 60ºC for 30 seconds.
Expected products
The high semolina color from cultivar Kofa produces a band of 200-bp, while low-color lines, like Desert King (DK) and UC1113, yields a product of 217-bp. The image below shows PCR products separated on a 6% polyacrylamide gel, following the steps detailed in Ref. 9. Lanes 1 and 3 correspond to heterozygous lines, their top band is the hybrid double stranded DNA that runs slowly due to conformation polymorphism.
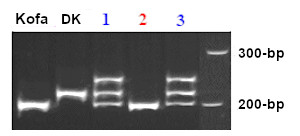
To get faster results, the PCR products can also be separated on a 2.5% agarose gel, but taking care of not loading wells with too much DNA. Results of a typical gel are shown below:

PSY-A1 primers (contributed by Carlos Guzmán García)
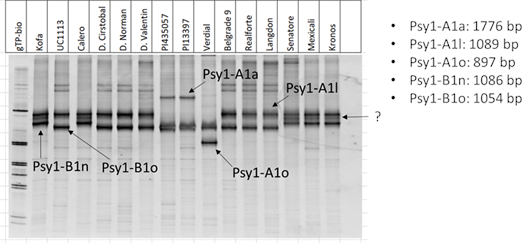
Singh et al. (13) developed a series of markers to study the allelic variation at the Psy1-A1 locus. One of these markers, Psy1-A1_STS, also differentiates some Psy1-B1 variants.
Psy1-A1_STS-F 5'- GTGGATATTCCCTGTCAGCATC -3'
Psy1-A1_STS-R 5'- GCCTCCTCGAAGAACATCCTC -3'
PCR mix:
DNA 50 ng
5X Buffer 1x
MgCl2 1.5 mM
dNTPs 0.2 mM
Primer-F 0.4 µM
Primer-R 0.4 µM
Taq Polymerase 1U
Water to 20 µl
PCR conditions:
- Denaturing step: 94°C, 5 min
- 35 cycles of: [94°C 30 sec, 56°C 30 sec, 72°C 60 sec]
- Extension step: 72°C, 10 min
Expected products
PCR products were separated on an 8% polyacrylamide gel.
Primers designed for scoring Lr19 and Sr25
Prins et al. developed a PCR marker, designated Gb, for the 7EL segment containing Y and Lr19 (10). The co-dominant marker, BF145935 derived from a wheat EST (11) was validated by Liu et al.(12) using 42 wheat lines and indicated that two DNA fragments amplified in most lines that differentiates Sr25 and non-Sr25 lines.
Note: you can find more details about the validation of these markers in Sr25 lines here.
The germplasm used by Prins et al. (10) was an allosynthetic recombinant produced several years earlier from the original translocation on 7D. An intercalary piece of the translocation was retained that only had Lr19 on it; both Y and Sr25 were lost during recombination, making it a suitable line for breeding in hexaploid lines because the white color of flour would be unaffected. Also the shorter translocation was relocated to 7BL during pairing induction. In spite of these rearrangements, this marker also detects the original Lr19 translocation on 7D (Frans Marais, personal communication).
Gb primer sequences
Gb-F 5'- CAT CCT TGG GGA CCT C -3’
Gb-R 5'- CCA GCT CGC ATA CAT CCA -3’
PCR conditions for Gb (times for each step might vary according to the thermocycler model):
- Denaturing step: 5 minutes at 94°C.
- Amplification step (40 cycles)
- 94°C, 30 sec
- 60°C, 30 sec
- 72°C, 1 min
- Extension step: 5 minutes at 72°C.
Final concentrations of the reagents used in the PCR amplification of Gb.
- 50-100 ng template DNA
- 5 pmol each primer
- 1 unit of Taq DNA polymerase
- 2 µl 15 mM MgCl2
- 2 µl 10X buffer
- 2 µl 1mM dNTPs
Total volume: 25 µl reaction
Expected products
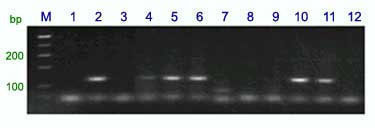
Following amplification for Gb, the products are separated in 2% agarose gels at 65 V constant voltage. The diagnostic fragment is 130-bp in length.
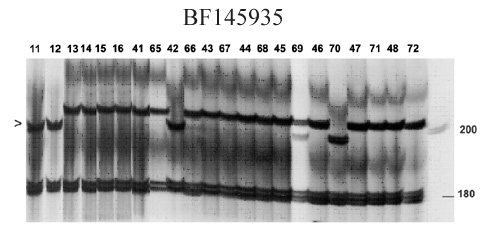
BF145935
The PCR marker based on EST BF145935 was originally designed for stem rust resistance gene Sr25, and you can find more details about these gene and marker validation here.
BF145935-F 5'- CTT CAC CTC CAA GGA GTT CCA C -3’
BF145935-R 5'- GCG TAC CTG ATC ACC ACC TTG AAG G -3’
PCR conditions for BF145935 (times for each step might vary according to the thermocycler model):
- Denaturing step: 94°C, 4 min
- Amplification step (35 cycles):
- 94°C, 45 sec
- 50°C, 30 sec
- 72°C, 45 sec
- Extension step: 72°C, 7 min
Expected products
Marker BF145935 produces fragment sizes of 198 and 180 bp in Sr25 lines and 202 and 180 bp in wheat lines without Sr25 (sse figure below).
References
1. Spectrophotometric determination of yellow pigment content and evaluation of carotenoids by high-performance liquid chromatography in durum wheat grain. Hentschel V, Kranl K, Hollmann J, Lindhauer MG, Bohm V, Bitsch R. In: Journal of Agricultural & Food Chemistry, 2002, 50(23):6663-6668. DOI:10.1021/jf025701p
2. Identification of a microsatellite on chromosome 7B showing a strong linkage with yellow pigment in durum wheat (Triticum turgidum L. var. durum). Elouafi I, Nachit MM, Martin LM, In: Hereditas, 2001, 135(2-3):255-261. DOI:10.1111/j.1601-5223.2001.t01-1-00255.x
3. Association of a lipoxygenase locus, Lpx-B1, with variation in lipoxygenase activity in durum wheat seeds. Hessler TG, Thomson MJ, Benscher D, Nachit MM, Sorrells ME. In: Crop Science, 2002, 42(5):1695-1700, 2002. DOI:10.2135/cropsci2002.1695
4. Mapping Loci Associated with Flour Colour in Wheat (Triticum Aestivum L.) . Parker GD, Chalmers KJ, Rathjen AJ, Langridge P. In: Theoretical & Applied Genetics, 1998, 97(1-2):238-245. DOI:10.1007/s001220050891
5. Development of a STS marker linked to a major locus controlling flour colour in wheat (Triticum aestivum L.). Parker GD, Langridge P. In: Molecular Breeding, 2000, 6(2):169-174. DOI:10.1023/A:1009638017859
6. Mapping components of flour and noodle colour in Australian wheat. Mares DJ, Campbell AW. In: Australian Journal of Agricultural Research, 2001, 52(11-12):1297-1309. DOI:10.1071/AR01048
7. Identification of QTL and association of a Phytoene synthase gene with endosperm colour in durum wheat. Pozniak CJ, Knox RE, Clarke FR, Clarke JM. Theorerical and Applied Genetics, 2007, 114:525-537. DOI:10.1007/s00122-006-0453-5
8. Molecular characterization of durum and common wheat recombinant lines carrying leaf rust resistance (Lr19) and yellow pigment (Y) genes from Lophopyrum ponticum. Zhang W, Soria MA, Goyal S, Dubcovsky J, Lukaszewski AJ, Kolmer J. In: Theoretical and Applied Genetics, 2005, 111: 573-582. DOI:10.1007/s00122-005-2048-y
9. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Zhang W, Dubcovsky J. In: Theoretical and Applied Genetics, 2008, 116.5 (2008): 635-645. DOI:10.1007/s00122-007-0697-8
10. AFLP and STS tagging of Lr19, a gene conferring resistance to leaf rust in wheat. Prins R, Groenewald JZ, Marais GF, Snape JW, Koebner RMD. In: Theoretical & Applied Genetics, 2001, 103(4):618-624. DOI:10.1007/PL00002918
11. Trigenomic chromosomes by recombination of Thinopyrum intermedium and Th. ponticum translocations in wheat.Ayala-Navarrete L, Bariana HS, Singh RP, Gibson JM, Mechanicos AA, Larkin PJ. In: Theoretical and Applied Genetics, 2007, 116:63-75. DOI:10.1007/s00122-007-0647-5.
12. Diagnostic and co-dominant PCR markers for wheat stem rust resistance genes Sr25 and Sr26. Liu S, Yu L-X, Singh RP, Jin Y, Sorrells ME, Anderson JA. In: Theoretical and Applied Genetics, 2010, 120:691–697. DOI:10.1007/s00122-009-1186-z.
13. Allelic variation at Psy1-A1 and association with yellow pigment in durum wheat grain. Singh A, Reimer S, Pozniak CJ, Clarke FR, Clarke JM, Knox RE, Singh, AK. In: Theoretical and Applied Genetics, 2009, 118:1539-1548. DOI:10.1007/s00122-009-1001-x.
