Contributed by Marcelo A. Soria, Wenjun Zhang and Jorge Dubcovsky. KASP marker contributed by Gina Brown-Guedira. We also acknowledge valuable contributions by Harbans Bariana, Manilal William, Beat Keller, Evans Lagudah and Bob McIntosh.
More than 60 leaf rust resistance genes and QTLs have been described in wheat (1). Many of them are race-specific genes, and several are currently used by breeders to develop new cultivars. However, the resistance provided by these genes can be short-lived as new races of the leaf rust pathogen, Puccinia triticina, are continuously evolving and acquire virulence against these genes. In order to obtain cultivars with good levels of protection under high disease pressure, several ‘slow rusting” gene complexes need to be combined. Besides, the effect of slow rusting genes is more dependent on environmental conditions. Wheat breeders can use slow rusting genes as a complement to race-specific genes. The availability of molecular markers greatly facilitates the pyramidization process.
A small group of leaf rust resistance genes are known as "slow rusting genes", such as Lr34 (2) and Lr46 (3). They provide durable and non-specific adult plant resistance but their effect is more reduced than that of race-specific genes. Lr34 was recently cloned (11) and it was shown that it is the same gene as Yr18 (resistance to adult plant stripe rust), powdery mildew resistance (Pm38) and leaf tip necrosis (Ltn1). This gene codes for a putative ABC transporter. A similar tight linkage or pleiotropic effect was observed between Lr46and Yr29, also a slow rusting gene for stripe rust.
Lr34 is located on the short arm of chromosome 7D, close to locus Xgwm295. The resistance phenotype displayed by this gene includes longer latent period, fewer uredina and smaller uredina size. Lr34 is tightly linked to the leaf tip necrosis (LTN) locus, it is also possible that the LTN phenotype could be a pleiotropic effect of Lr34 itself (5).
Markers for Lr34-Yr18-Sr57
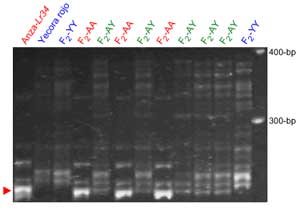
For some CIMMYT germplasms the LTN phenotype can be used directly as a morphological marker (6). Lagudah et al (12) developed an STS marker, csLV34, that maps 0.4 cM from Lr34, and was validated in many lines and cultivars from different breeding programs worldwide. In the paper reporting the cloning of Lr34 (11), Krattinger et al. described several markers that differentiate among the loss-of-function alleles of Lr34. In addition, a KASP marker is available.
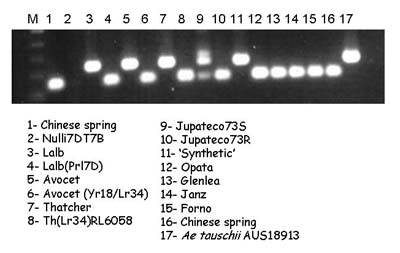
STS marker csLV34
Primer sequences:
csLV34F 5'- GTT GGT TAA GAC TGG TGA TGG -3'
csLV34R 5'- TGC TTG CTA TTG CTG AAT AGT -3'
PCR conditions:
- Denaturing step: 94°C, 5 min
- Amplification step (40 cycles):
- 94°C, 45 s
- 55°C, 30 s
- 72°C, 60 s
- Extension step: 72°C, 7 min
Expected products:
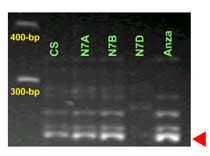
The Lr34 allele yields a 150-bp product, and a 229-bp band is amplified in non-Lr34 germplasm
KASP marker (contributed by Gina Brown-Guedira)
Marker wMAS000003 is used to identify the presence of Lr34 and is designed around a 3 bp indel in exon 11 of the Lr34/Yr18 gene. Lines testing positive (VIC allele) for Lr34 also need to be tested with the Lr34 Jagger assay which identifies the nonfunctional allele of Lr34 from Jagger. This assay is based on a mutation in exon 22 of the resistance gene (Lagudah et al., 2009). Lines that are positive for Lr34 using the wMAS000003 assay and exhibit the FAM allele with the Lr34 Jagger assay are considered to have a functional Lr34 allele whereas lines that are positive for Lr34 using the wMAS000003 assay and exhibit the VIC allele with the Lr34 Jagger assay are considered to have a nonfunctional Lr34 Jagger allele.
KASP marker (contributed by Gina Brown-Guedira)
Marker wMAS000003 is used to identify the presence of Lr34 and is designed around a 3 bp indel in exon 11 of the Lr34/Yr18 gene. Lines testing positive (VIC allele) for Lr34 also need to be tested with the Lr34 Jagger assay which identifies the nonfunctional allele of Lr34 from Jagger. This assay is based on a mutation in exon 22 of the resistance gene (13). Lines that are positive for Lr34 using the wMAS000003 assay and exhibit the FAM allele with the Lr34 Jagger assay are considered to have a functional Lr34 allele whereas lines that are positive for Lr34 using the wMAS000003 assay and exhibit the VIC allele with the Lr34 Jagger assay are considered to have a nonfunctional Lr34 Jagger allele.
|
|
wMAS000003 |
Lr34 Jagger |
|
lr34 |
FAM allele |
FAM allele |
|
Lr34 |
VIC allele |
FAM allele |
|
Lr34 Jagger |
VIC allele |
VIC allele |
The KASP assays (KBiosciences competitive allele-specific PCR) were performed according to manufacturer’s instructions, the endpoint genotyping was performed on a PHERAstar Plus microplate reader and data were analyzed using the KlusterCaller software.
|
SNP ID |
wMAS000003 |
not applicable |
|
Gene |
Lr34 |
Lr34 - Jagger |
|
Evidence |
causal |
causal |
|
Primer Allele FAM |
CTGGTATGCCATTTAACATAATCATGAA |
TGTAATGTATCGTGAGAGATTTGCAG |
|
Primer Allele VIC |
CTGGTATGCCATTTAACATAATCATGAT |
ATTGTAATGTATCGTGAGAGATTTGCAT |
|
Primer Common |
CGCATGACAATAAGTTTCACTCATGCAAA |
GATCATTATCTGACCTGTGCGAATGAATA |
|
FAM allele |
A |
G |
|
PAM allele |
T |
T |
|
FAM phenotype |
susceptible |
susceptible / resistant |
|
VIC phenotype |
resistant |
susceptible |
Other markers
Previously to the development of STS marker csLV34 and the positional cloning of Lr34, there were several markers available that mapped farther away and were validated in some populations. Suenaga et al. (7) had determined that Xgwm295 and Xgwm130 were closely linked to each other at the center of the slow rusting QTL. Later, Schnurbusch et al. (10) analyzing an Arina x Forno population found that the region containing Lr34 and flanked by Xbarc352 (distal) and Xgwm295 (proximal) spanned 17.7 cM. Below we provide protocols for using this markers.
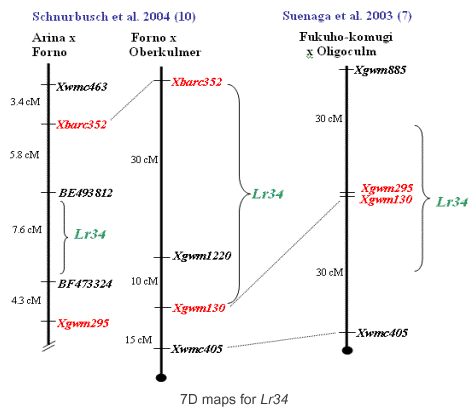
Microsatellite marker Xgwm295
The microsatellite locus Xgwm295 maps close to the Lr34 locus but its detection is sometimes difficult to optimize. Details of the PCR protocol can be obtained from the WMS295 probe record in GrainGenes.
Manilal William, personal communication: the Xgwm295 allele putatively linked to Lr34/Yr18 has a size of approximately 250-bp.
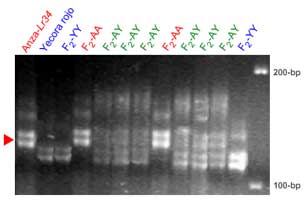
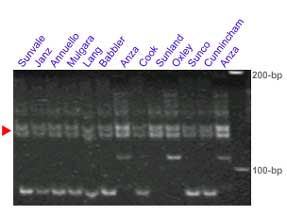
Microsatellite markers Xgwm130 and Xbarc352
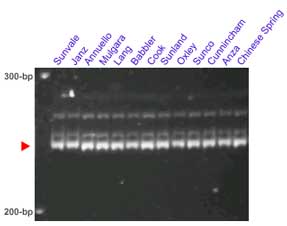
The microsatellite loci Xgwm130 and Xbarc352 flank the Lr34 locus. In some crosses these set of markers provides clearer results than Xgwm295. This pair of markers was tested in several lines carrying Lr34: Sunvale, Janz, Annuello, Mulgara, Lang, Babbler, Cook, Sunland, Oxley, Sunco, Cunnincham and Anza,
Primer sequences:
Xgwm130:
WMS130-L 5'- AGC TCT GCT TCA CGA GGA AG -3'
WMS130-R 5'- CTC CTC TTT ATA TCG CGT CCC -3'
Xbarc352:
barc352-L 5'- CCC TTT CTC GCT CGC CTA TCC C -3'
barc352-R 5'- CTG TTT CGC CCA ATC TCG GTG TG -3'
Final concentrations of the reagents used in the PCR amplification:
- Genomic DNA: 50 ng
- Primers: 5 pM each
- dNTPs: 100 µM each
- Taq DNA polymerase: 1U
- 10x PCR buffer
- 1.5 mM MgCl2
Total volume: 20 µl
PCR conditions:
Xgwm130:
- Denaturing step: 94°C, 5 min
- Amplification step (45 cycles):
- 94°C, 20 s
- 58°C, 20 s
- 72°C, 30 s
- Extension step: 72°C, 7 min
Xbarc352:
- Denaturing step: 94°C, 4 min
- Amplification step (38 cycles):
- 94°C, 30 s
- 60°C, 30 s
- 72°C, 60 s
- Extension step: 72°C, 5 min
Expected products:
All the gels shown below are 6% polyacryalmide gels stained with ethidium bromide and visualized under UV light (method).
Xgwm130:
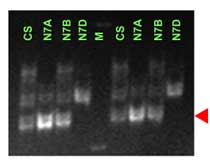
Xgwm130 is in a proximal position relative to Lr34 (ref. 10 and also see maps in the main section). One potential problem with this marker is that it detects more than one locus. The red arrowhead indicates a putative identification of the band cosegregating with Lr34. The information provided should be considered preliminary.
Please read carefully the communications below from people also working with this marker. Input from users is welcome.
R.Rahaman, H.Bariana, M.Shriflou and P. Sharp personal communication: gwm130 showed a band of 133-bp in 21 genotypes carrying Lr34, whereas a band of 135-bp is amplified in 20 genotypes lacking Lr34. The Lr34 carrying cultivar Suneca was an exception, where a 120 bp-band was amplified.
Manilal William, personal communication: In some populations (Fukokumughi x Oligoculm and Avocet x Parula) the Xgwm130 allele is scored as a codominant band, while in other populations (Frontana x Inia66) it has been scored as a dominant band, which is present in Inia66 and absent in Frontana carrying Lr34/Yr18). The apparent size of the +/- allele is about 110. In multiple mapping populations it was not possible to map any allele of Xgwm130 on 7A.
Xbarc352:
Available germplasm
Lr34 is a gene present in many parent lines that have been used for breeding commercial varietes around the world. The STS marker csLV34 was proved an effective marker to differentiate between llines carrying Lr34 and its isogenic counterparts without the gene. Among others, csLV34 was validated in Thatcher, Glenlea, Jupateco R, Opata, Fukuho-komugi, Condor, Cook, Anza, Forno, Bezostaya, Otane and Chinese Spring.
Additional information. Field testing
Singh and Huerta Espino (8) analyzed different yield-related parameters in the Mexican spring cultivar Jupateco and a near isogenic line (NIL) derived from it and carrying Lr34 (Jutapeco-Lr34). They found that in disease-free environments Jupateco-Lr34 had significant, but generally small, reductions in biomass, grain per spike and grains/m2. In some experiments they also observed negative effects of Lr34 on mean grain yield and mean grain weight. On the other hand, in leaf-rust affected plots Jutapeco showed significantly higher yield penalties than Jupateco-Lr34.
Labuschagne et al. (9) studied the effect of Lr34 on breadmaking quality in the South African cultivar Kareen. They compared Kareen with several different NILs derived from it and carrying Lr34. Kareen-Lr34 NILs had a flour protein content and water absorption values significantly higher than that of Kareen, while flour extraction and mixograph development time were similar. Only one Lr34-NIL showed a significantly lower SDS-sedimentation value. The authors concluded that Lr34 has an overall advantageous effect on breadmaking quality on this line.
References
1. Catalogue of gene symbols for wheat. McIntosh RA, Yamazaki Y, .Devos KM, Dubcovsky J, Rogers R, Appels R. In: KOMUGI, Integrated wheat Science Database
2. Effect of leaf rust resistance gene Lr34 on components of slow rusting at seven growth stages in wheat. Singh RP, Huerta-Espino J. In: Euphytica, 2003, 129:371-376.
3. Characterization of Lr46, a gene conferring partial resistance to wheat leaf rust. Martínez F, Niks RE, Singh RP, Rubiales D. In: Hereditas, 2001, 135:111-114.
4. Inheritance of adult-plant leaf rust resistance derived from the common wheat varieties Exchange and Frontana. Dyck PL, Samborski DJ, Anderson RG. In: Canadian Journal of Genetics and Cytology, 1966, 8:665-671.
5. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Schnurbusch T, Paillard S, Schori A, Messmer M, Schachermayr G, Winzeler M, Keller B. In: Theoretical and Applied Genetics, 2004, 108:477-484.
6. Association between gene Lr34 for leaf rust resistance and leaf tip necrosis in wheat. Singh RP. In: Crop Science, 1992, 32: 874-878.
7. Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Suenaga K, Singh RP, Huerta-Espino J, William HM. In: Phytopathology, 2003, 93:881-890.
8. Effect of leaf rust resistance gene Lr34 on grain yield and agronomic traits of spring wheat. Singh RP, Huerta-Espino J. In: Crop-Science, 1997, 37:390-395.
9. The influence of leaf rust resistance genes Lr29, Lr34, Lr35 and Lr37 on breadmaking quality in wheat. Labuschagne MT, Pretorius ZA, Grobbelaar B. In: Euphytica,2002, 124: 65–70.
10. Tagging and validation of a major quantitative trait locus for leaf rust resistance and leaf tip necrosis in winter wheat cultivar forno. Schnurbusch T, Bossolini E, Messmer B, Keller B.In: Phytopathology, 2004, 94: 1036-1041.
11. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J,McFadden H, Bossolini E, Selter LL, Keller B. In: Science, 2009, 323:1360-1363. DOI: 10.1126/science.1166453.
12. Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Lagudah ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, Spielmeyer W. In: TAG Theoretical and Applied Genetics, 2006, 114:21-30. DOI: 10.1007/s00122-006-0406-z.
13. Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, Brown-Guedira G, Selter LL, Keller B. In: Theoretical and Applied Genetics, 2009, 119:889-898. DOI:10.1007/s00122-009-1097-z.
