The rice Grain Width and Weight 2 (OsGW2) gene was previously described as a negative regulator of grain weight and width. It encodes a RING-type E3 ubiquitin ligase (1). The wheat homolog in wheat is the TaGW2 gene, with homoeologues in the three genomes, although the most closely studied to date is the A genome homoeologue, TaGW2-A1. Two studies showed in Chinese wheat lines a significant association of a single nucleotide polymorphism (SNP) in the promoter region of TaGW2-A1 with thousand grain weight (TGW) and grain width (2, 3). These studies also showed a negative relationship between transcript levels and grain width. Another study determined that a single base insertion in the last exon of TaGW2-A1 produced a truncated protein that was associated with increases in TGW and grain width (4). In contrast, Bednarek et al. (5) down-regulated the expression of the threeTaGW2 homoeologues by RNAi and found decreases in grain weight and size in hexaploid wheat. It cannot be ruled out that TaGW2 has additional functions, and the drastic reduction in expression levels seen in the RNAi experiments could affect those functions in a different way than the absence of only one of the homoeologues.
Simmonds et al (6) screened the tetraploid Kronos TILLING population to identify mutants in TaGW2-A1. They identified a G to A transition, designated G2373A, in the splice acceptor site of exon 5 which leads to two variants of mis-splicing inTaGW2-A1. The most frequent variant results in a frame-shift that generates a premature termination codon that yields a shorter protein of 134 amino acids. The other is an in-frame variant missing three amino acids. The mutant allele was introgressed into tetraploid and hexaploid germplasm and tested in thirteen greenhouse and field experiments. Overall the TaGW2-A1 mutant allele significantly increased TWG (6.6%), grain width (2.8%) and grain length (2.1%). The authors developed a KASP assay to discriminate between the wildtype and mutant in the splice acceptor site of exon 5.
Two tetraploid lines wheat lines carrying the mutant allele are available to wheat breeders. One is a KRONOS isogenic line (4X) and the other is a Paragon isogenic line (6X). JIC Germplasm Resources Unit (http://www.seedstor.ac.uk/) accession numbers: W10281-W10284; US National Small Grains Collection accession numbers: PI 675010, PI 675011, PI 675012, PI 675013, PI 675014, PI 675015.
We provide below protocols for KASP markers and a CAPS marker to score TaGW2-A1.
KASP Marker for TaGW2-A1
Simmonds et al (6) developed a KASP assay to differentiate the two variants of the G2373A SNP. Two allele specific reverse primers were designed with either a G or A at the 3' end: primer K2 for the wildtype (G) and K3 for the mutant (A) allele. The common primer (K1) includes a 3-bp indel present in the A genome and absent in the B genome. The allele specific primers (K2 and K3) carry FAM and HEX tails.
Primer sequences:
K1_TaGW2_Aspecific 5'- AGA GCA ATT TGT AAG TCT TAT TCC -3'
K2_TaGW2_Awt 5'- GCT TCA ATG ACT TTC TGT TCT TCC -3'
K3_TaGW2_Amutant 5'- GCT TCA ATG ACT TTC TGT TCT TCT -3'

Primer mix
- 46 µl deionized water
- 30 µl common primer (100 µM)
- 12 µl of each tailed primer (100 µM)
PCR mix
- 2.50 µl template (10 - 20 ng of DNA)
- 2.43 µl V3 2X Kaspar mix
- 0.07 primer mix
PCR conditions
- Hotstart at 95°C for 15 min
- Touchdown (10 cycles):
- 95°C, 20 sec
- 65°C, -1 °C per cycle, 25 sec
- 72°C, 45 sec
- Amplification (26 cycles):
- 95°C, 10 sec
- 57°C, 60 sec
Since the amplicon size is small, 121-bp, there is no need for extension steps. PCR was performed on a Peltier PTC-225. Fluorescent end-point genotyping was carried out using a Tecan Safire plate reader (Tecan Group AG, Mannerdorf, Switzerland). Two images of typical results obtained with tetraploid (Kronos) and hexaploid lines (Paragon) and their respective mutants and heterozygous are shown below.
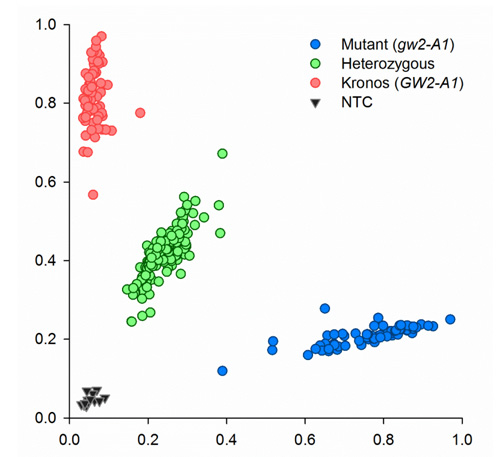
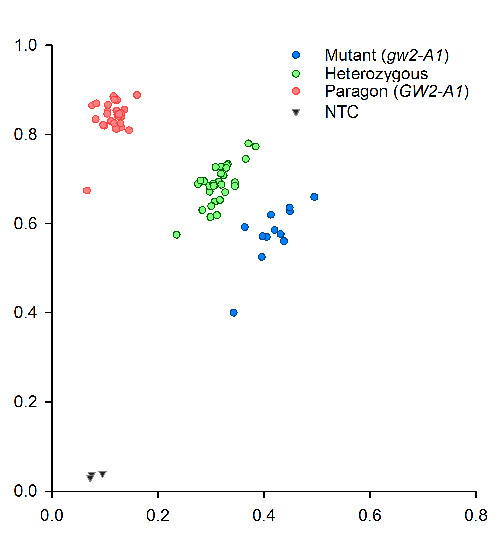
CAPS marker
This marker requires a PCR amplification with the primer pair F1/R2 followed by a digestion with BsrI.
Primer sequences:
GW2A_dCAP_F1 5'- ACTTTATCTTGTATAAATTTTGTTACT -3'
GW2A_dCAP_R2 5'- TAGTAAATTGTAAGTTATCAGATTAGA -3'
PCR mix
PCR buffer: 10 mM Tris–HCl, PH 8.8; 50 mM KCl; 0.01% Tween 20
1.5 mM MgCl2
0.1 mM dNTPs
0.5 μM of each primer
50–100 ng of genomic DNA
1 U Taq DNA polymerase
Digestion mix
0.35 U of BsrI
Neb3.1 buffer:
100 mM NaCl
50 mM Tris-HCl
10 mM MgCl2
100 µg/ml BSA
pH 7.9@25°C
PCR and digestion conditions:
Annealing temperature: 50°C,
Extension time = 45 secs.
40 cycles
Digest with BsrI
Expected products
The wild-type allele yields two products of 30 and 241 bp. The mutant allele yields three bands of 30, 241 and 271 bp. An image of digestion products separated on a 6% gel is shown:
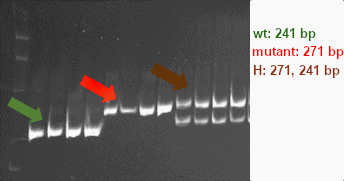
References
1. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Song X-J, Huang W, Shi M, Zhu M-Z, Lin H-X. In: Nature Genetics, 2007, 39:623-630. DOI:10.1038/ng2014.
2. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Su Z, Hao C, Wang L, Dong Y, Zhang X In: Theoretical and Applied Genetics, 2011, 122:211-223. DOI:10.1007/s00122-010-1437-z
3. Function of TaGW2-6A and its effect on grain weight in wheat (Triticum aestivum L.). Zhang X, Chen J, Shi C, Chen J, Zheng F, Tian J In: Euphytica, 2013, 192:347-357. DOI:10.1007/s10681-012-0858-y.
4. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Yang Z, Bai Z, Li X, Wang P, Wu Q, Yang L, Li L, Li X In: Theoretical and Applied Genetics, 2012, 125:1057-1068. DOI:10.1007/s00122-012-1895-6.
5. Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. Bednarek J, Boulaflous A, Girousse C, Ravel C, Tassy C, Barret P, Bouzidi MF, Mouzeyar S In: Journal of Experimental Botany, 2012, 63:5945-5955. DOI:10.1093/jxb/ers249.
6. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Simmonds J, Scott P, Brinton J, Mestre TC, Bush M, del Blanco A, Dubcovsky J, Uauy C. In: Theoretical and Applied Genetics, 2016, 129:1099-1112. DOI:10.1007/s00122-016-2686-2.
