In 1970, Gerechter-Amitai and Stubbs (1) reported that accession G-25 of Triticum dicoccoides Korn was resistant to many races of Puccinia striiformis from different geographical origins. Later, it was shown that this stripe rust resistance was conferred by the dominant gene Yr15 (2,3). McIntosh and Silk showed that it is located on the short arm of chromosome 1B (4). Yr15 was introgressed into tetraploid and hexaploid wheats (2, 5). Xianming Chen informed that after several years of greenhouse and field testing, no virulent isolates on Yr15 have been detected (Xianming Chen, personal communication).
In 2018 Klymiuk et al. (7) reported the cloning of Yr15. They found that Yr15 encodes a protein composed of putative kinase and pseudokinase domains in tandem and it was designated WTK1 (wheat tandem kinase 1). Further comparative sequence analysis revealed that Yr15 is a member of a novel family of plant proteins named tandem kinase proteins (TKPs, 8).
Markers for Yr15
There are different molecular markers available for Yr15. Different breeding programs have found that some of the markers are not polymorphic for their populations. However, there are several markers close to Yr15 that can be tested. Peng et. al (6) published a high density map of the chromosome region where Yr15 is located (see fig. 2 of that publication for a map and distances). More recently a series of close CAPS markers and perfect KASP markers were developed.
SSR markers for Yr15
| Microsatellite | Position relative to Yr15 (based on reference 6) |
|---|---|
| Proximal loci | |
| gwm413 | 4.4 cM |
| gwm273 | 5.7 cM |
| gwm11 | 6.2 cM |
| gwm18 | 6.9 cM |
| Distal loci | |
| barc8 | 9 cM distal. Based on BARC map |
| gwm911 | 21 cM |
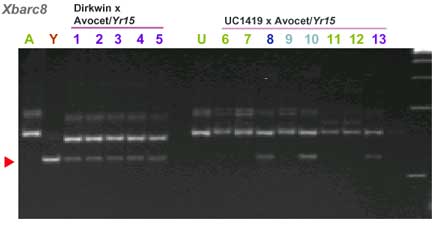
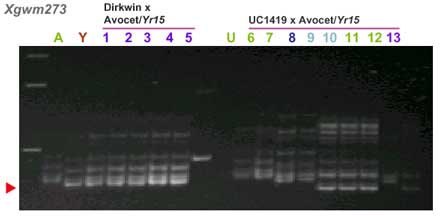
CAPS markers for Yr15 (ref. 7)
uhw296
The codominant marker uhw296 was confirmed to co-segregate with Wtk1 (Yr15) or with a close proximal location, depending on the population.
Primers for uhw296:
uhw296_F: CAACCGTGCCTCCAAACA
uhw296_R: CGGGTGTTGTCCGTTGAG.
PCR and digestion conditions:
- Touchdown: 65ºC/55ºC, with 50 sec elongation time.
- Digest with NlaIV restriction enzyme.
- Expected products:
- B9 (Yr15 line): 321, 176
- D447 (susceptible line): 299, 176, 22.
uhw273
The next closest proximal codominant marker is the uhw273 CAPS
Primers for uhw273:
uhw273_F: GGTGACGGCGAGTGTACG
uhw273_R: GACGCAATTGTCCGCTGT
PCR and digestion conditions:
- Touchdown: 62ºC/52ºC, with 25 sec elongation time.
- Digest with EagI restriction enzyme.
- Expected products:
- B9 (Yr15 line): 149, 93
- D447 (susceptible line): 242
uhw297
The closest distal codominant marker is the uhw297.
Primers for uhw297:
uhw297_F: CAGATGACCAACCAAAAGCA
uhw297_R: GTCATATTGGTGCCCAGTGA
PCR and digestion conditions:
- Touchdown: 65ºC/55ºC, with 40 sec elongation time.
- Digest with BauI restriction enzyme.
- Expected products:
- B9 (Yr15 line): 487
- D447 (susceptible line): 450, 37
KASP markers for Yr15 (ref. 9)
There are two perfect, co-dominant KASP markers, Kin1 and Kin2 that target the KinI and KinII domains, respectively. These primers are intellectual property of Carmel, the economic corporation of Haifa University, so if you plan to use them for commercial applications, read information below:
Please note that the information presented (including all rights thereto) is intellectual property owned by Carmel – The economic corporation of Haifa University and/or its third party licensors which is, inter alia, protected under patent applications. If you wish to make any use of such information you are required to contact Carmel and obtain its prior written approval at carmel@univ.haifa.ac.il
Primers for Kin1
Kin1_A: GACCCTTTGTTTTGTCAGAGC
Kin1_B: GACCCTTTGTTTTGTCAGAGT
Kin1_C (common): AAACATTGAGTAAGACTAGTAGCC
Primers for Kin2
Kin2_A: CTGCTACTGGACAGTGTAACG
Kin2_B: CTGCTACTGGACAGTGCAACA
Kin2_C (common): CCCATGTCACGAGGCT
The StepOne Plus Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA) was used for amplification under the following conditions: TD – 61°C, 60 sec (15 cycles) / 55°C, 60 sec (32 cycles)
Although these markers are co-dominant, the Wtk1 functional allele does not look like homozygous allele on the plot. This is because it amplifies as well other WTK1 copies from other than the 1BS location (see Fig. S2 of Ref. 9).
References
1. A valuable source of yellow rust resistance in Israeli population of wild emmer Triticum dicoccoides Koern. Gerechter-Amitai ZK, Stubbs RW. In: Euphytica, 1970, 19:12-21.
2. Inheritance of resistance to stripe rust (Puccinia striiformmis) in crosses between wild emmer (Triticum dicoccoides) and cultivated tetraploid and hexaploid wheat. I. Triticum durum. Gerechter-Amitai ZK, Grama A. In: Euphytica, 1974. 23:387-392.
3. Yr15: A new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G-25. Gerechter-Amitai ZK, Van Silfhout CH, Grama A, Kleitman F. In: Euphytica, 1989, 43(1-2):187-190.
4. Cytogenetic studies in wheat XVII. Monosomic analysis and linkage relationships of gene Yr15 for resistance to stripe rust. McIntosh RA, Silk J, The TT. In: Euphytica, 1996, 89(3):395-399.
5. Inheritance of resistance to stripe rust (Puccinia striiformmis) in crosses between wild emmer (Triticum dicoccoides) and cultivated tetraploid and hexaploid wheat. I. Grama A, Gerechter-Amitai ZK. In: Euphytica, 1974. 23:393-398.
6. High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides.. Peng JH, Fahima T, Roeder MS, Huang QY. Dahan A, Li YC, Grama A, Nevo E. In: Genetica, 2000, 109(3): 199-210.
7. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Klymiuk, V, Yaniv E, Huang L, Raats D, Fatiukha A, Chen S, Feng L, Frenkel Z, Krugman T, Lidzbarsky G, Chang W, Jääskeläinen MJ, Schudoma C, Paulin L, Laine P, Bariana H, Sela H, Saleem K, Sørensen CK, … Fahima T. (2018). Nature Communications, 2018 9(1), 1–12. DOI: 10.1038/s41467-018-06138-9
8. Tandem protein kinases emerge as new regulators of plant immunity. Klymiuk V., Coaker G., Fahima T., Pozniak C.J. (2021) . Molecular Plant-Microbe Interactions, 2021. DOI:10.1094/MPMI-03-21-0073-CR.
9. Wheat tandem kinases provide insights on disease resistance gene flow and host-parasite co-evolution. Klymiuk V, Fatiukha A, Fahima T. The Plant Journal, 2019, 98, 667-679 DOI:10.1111/tpj.14264.
