The leaf rust resistance gene Lr47 confers resistance to a wide spectrum of leaf rust strains. This gene was transferred from chromosome 7S of Triticum speltoides to chromosome 7A of Triticum aestivum. Initially, the RFLP locus Xabc465 was used to follow the transfer of the segment containing Lr47 (1), but later, this marker was converted into a dominant PCR marker. To detect the allele present in 7A, a cleavege amplified polymorphic sequence (CAPS) marker was developed. Using both markers, homozygous and heterozygous individuals for Lr47 can be discriminated.
Markers for Lr47
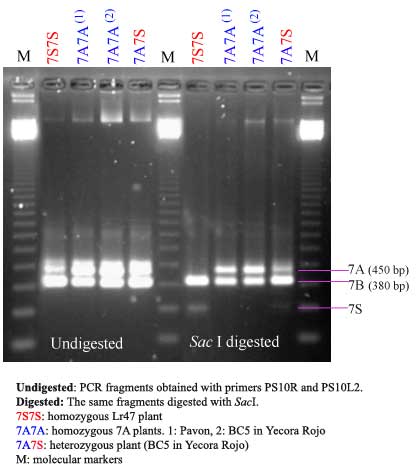
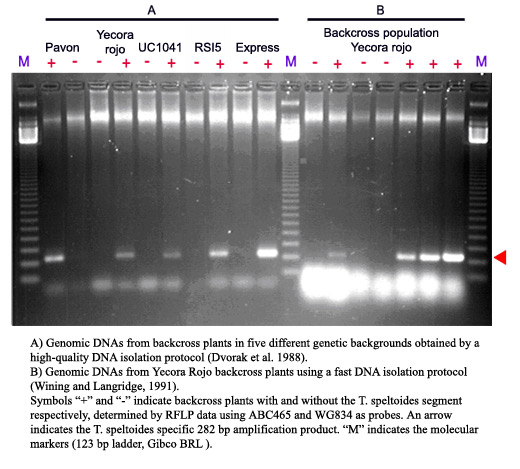
Since the T. speltoides segment containing Lr47 generally will not recombine in the presence of a wild-type Ph1, only one marker is needed to determine the presence of the whole segment. The primers specific for T. speltoides are PS10R and PS10L, which produce a band of 282 bp To differentiate homozygous from heterozygous Lr47 plants a CAPS marker was developed that is specific for the 7A allele of the Xabc465 locus. This marker is detected using primers PS10R and PS10L2 and then digesting with Sac I. See below for the laboratory protocol and Ref.2 for additional details.
CAPS-specific primers for the A genome allele to differentiate AS and SS genotypes
The PCR primers designed for detecting the 7S segment cannot be used to discriminate between heterozygous and homozygous individuals for this segment. In homozygous plants for the 7S segment the A genome allele of Xabc465 is absent, so a CAPS (Cleavage Amplified Polymorphic Sequence) marker was designed to detect this allele.
PCR Primers PS10R and PS10L2 are used to amplify a DNA fragment, the product obtained is digested with SacI and if the individual carries the A genome allele, a 450-bp product is observed. This product is absent in homozygous plants for 7S.
DNA extraction: RNAsa treatment is recommended to see the PCR product in agarose gels
Primers sequences:
PS10R 5’ GCT GAT GAC CCT GAC CGG T 3’
PS10L2 5’ GGG CAG GCG TTT ATT CCA G 3’
PCR amplification conditions:
- Denaturing step: 94° 3 min
- 40 cycles of: [94° 30 seconds, 55° 30 seconds, 72° 30 seconds]
- Extension step: 72° 7 min
Final volume: 25 µl
Final concentrations of the reagents used in the PCR reaction:
- 1X Taq DNA polymerase buffer (Promega)
- MgCl2: 3.0 mM
- primers: 0.2 µM each
- dNTP: 200 µM each
- DNA input: 120 ng/reaction
- Taq polymerase [5 U/ml]: 0.3 µl/reaction (1.5U/reaction)
SacI digestion (DNA precipitation may be necessary)
DNA 5.0 µl (half of PCR-precipitated product)
10X buffer B for Sac I 2.0 µl
BSA 0.2 µl
SacI [10U/µl] 0.5 µl
dd water 12.3 µl
Final vol. 20.0 µl (Incubate @ 37 ° during 2 hs)
Electrophoresis: Agarose 3% long gels, 2 hs and 30 min @ 80V
Expected undigested product size: 452 bp (A genome allele) and 400 bp (B genome allele).
Expected digested product size: 246 and 238 bp (S allele).
PCR-specific primers for the T. speltoides S genome allele of Xabc465
DNA extraction: RNAsa treatment is recommended to see the PCR products in agarose gels
Primers sequences:
PS10R: 5' GCT GAT GAC CCT GAC CGG T 3'
PS10L: 5' TCT TCA TGC CCG GTC GGG T 3'
PCR amplification conditions:
- Denaturing step: 94°, 3 min
- Touchdown from 70° to 64°. Seven cycles of: denaturation at 94 ° for 30 seconds, in every cycle the annealing temperature steps down {70, 69, 68, 67, 66, 65, 64 °C} for 30 seconds and extension at 72 ° for 30 seconds
- 35 cycles of: [94° 30 seconds, 63° 30 seconds, 72° 30 seconds]
- Extension step: 72° 7 min
Final volume: 25 µl
Final concentrations of the reagents used in the PCR reaction:
- 1X Taq DNA polymerase buffer (Promega)
- MgCl2: 3.0 mM
- primers: 0.2 µM each
- dNTP: 200 µM each
- DNA input: 120 ng/reaction
- Taq polymerase [5 U/ml]: 0.3 µl/reaction (1.5U/reaction)
Electrophoresis: Agarose 2% gels
Expected PCR product size: 282 bp
Alternative codominant PCR marker
Contributed by Marcelo Helguera (mhelguera@correo.inta.gov.ar)
A single PCR reaction including three primers at different concentrations can be used to generate a codominant marker for the Lr47 chromosome segment. It is recommended to adjust first the annealing temperature to each specific laboratory condition, by searching for the temperature that results in similar fragment intensities in a heterozygous individual (F1).
Primers sequences:
PCAPSR: 5'- CGT GAG ACT CGC CGT TAC CTT G -3'
PS10L: 5'- TCT TCA TGC CCG GTC GGG T -3'
PS10L2: 5'- GGG CAG GCG TTT ATT CCA G -3'
PCR amplification conditions:
- Denaturing step: 94° 3 min
- 40 cycles of: [94° 45 seconds, 61° 30 seconds, 72° 30 seconds]
- Extension step: 72° 7 min
Final volume: 25 µl
Final concentrations of the reagents used in the PCR reaction:
- 1X Taq DNA polymerase buffer (Promega)
- MgCl2: 1.5 mM
- Primers:
- PCAPSR 0.2 µM
- PS10L 0.4 µM
- PS10L2 0.1 µM.
- dNTP: 200 µM each
- DNA input: 120 ng/reaction
- Taq polymerase [5 U/ml]: 0.2 µl/reaction (1 U/reaction)
Electrophoresis: Separate 10 µl of the PCR product in 1.5% agarose gel (no digestion required).
Expected digested product size: 224 (S allele) and 394 bp (A Allele).
1. Molecular characterization of two Triticum speltoides interstitial translocations carrying leaf rust and greenbug resistance genes. Dubcovsky, J.; Lukaszewski, A. J.; Echaide, M.; Antonelli, E. F.; Porter, D. R. In: Crop Science Nov.-Dec., 1998. 38 (6): 1655-1660.
2. Development of PCR markers for wheat leaf rust resistance gene Lr47. Helguera, M.; Khan, I. A.; Dubcovsky, J. In:Theoretical and Applied Genetics September, 2000. 101 (4): 625-631.
3. Registration of two germplasms of common wheat with interstitial translocations of Triticum speltoides carrying leaf rust and greenbug resistance genes. Lukaszewski, A. J.; Porter, D.R.; Antonelli, E. F.; Dubcovsky, J.In: Crop Science Mar.-Apr., 2000. 40:590.
