Triticum speltoides Taush (2n = 14, S genome) is an attractive source of high levels of resistance to leaf, stem and stripe rust of wheat (1). Leaf rust resistance genes Lr28, Lr35, Lr36,and Lr47 were derived from this species (3). Crosses between T. speltoides and hexaploid wheat (T. aestivum L. 2n=42, ABD genomes) followed by selection of resistant progeny frequently resulted in translocations between the T. speltoides and common wheat chromosomes because many T. speltoides genotypes have the ability to promote homoeologous chromosome pairing when hybridized with wheat (1).
Leaf rust resistance gene Lr51, formerly known as LrF7, was transferred from Triticum speltoides to common wheat by J. Dvorak (1). Lr51 is located within a 15-30-cM segment of Triticum speltoides chromosome 1S translocated to the long arm of chromosome 1B of bread wheat (2). In tests for resistance to Puccinia triticina race 5, J. Dvorak showed that plants homozygous for Lr51 were highly resistant with hypersensitive flecks, whereas heterozygous plants showed slightly lower levels of resistance with small pustules surrounded by necrosis and chlorosis, indicating incomplete dominance (1). A more recent study (2) showed that Lr51 is resistant to the current predominant races of leaf rust (P triticina) in the U.S. (see table below).
Lr51 has not been frequently used for breeding, in spite of the good levels of resistance it confers. Probably because the presence of large T. speltoides chromosome segments in a wheat background was associated with likely negative effects.
Markers for Lr51
CAPS Marker for Lr51
Helguera et al. (2) developed a CAPS (Cleaveged Amplified Polymorphic Sequence) marker for the AGA7 gene located within the T. speltoides translocation containing Lr51. Primers S30-13L and AGA7-759R were designed to amplify preferentially AGA7 alleles from the S and B genomes, and to discriminate against the A and D genomes. Since the T. speltoides segment does not recombine with the wheat chromosomes in a Ph background, only one marker is enough to follow the segment in a breeding program.
Primers sequences:
S30-13L 5'- GCA TCA ACA AGA TAT TCG TTA TGA CC -3'
AGA7-759R 5'- TGG CTG CTC AGA AAA CTG GAC C -3'
Final concentrations of the reagents used in the PCR amplification
- 100 ng genomic DNA
- 0.2 µM each primer
- 200 µM each dNTP
- 1U Taq DNA polymerase (Promega)
- 1x Taq polymerase buffer (Promega)
- 1.5 mM MgCl2
Total volume: 25 µl
PCR conditions:
- Denaturing step: 94°C, 4 min
- Amplification step (40 cycles):
- 94°C, 45 s
- 52°C, 45 s
- 72°C, 60 s
- Extension step: 72°C, 10 min
Enzymatic digestion
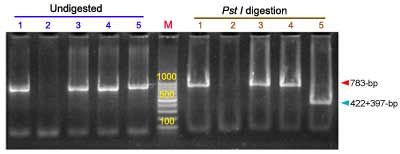
10 µl of the PCR amplification products were digested with restriction enzymes Pst I or BamH I (Promega) by adding 5 units of enzyme to the PCR product. The mix was incubated for 2 h at 37°C.
Expected products:
After digestion samples were separated by electrophoresis in 2% agarose gel and visualized using ethidium bromide and UV light. Pst I cut only the S genome amplification product (397-bp and 422-bp), whereas BamH I cut only the B genome amplification product (672-bp and 111-bp).
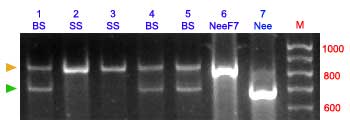
The amplification products from primers S30-13L /AGA7-759R from the B (783-bp) and S (819-bp) alleles can be also separated using polyacrylamide gels (14 %) without digestion.
References
1. Transfer of leaf rust resistance from Aegilops speltoides to Triticum aestivum. Dvorak J. In: Canadian Journal of Genetics and Cytology, 1977, 19:133-141. DOI: 10.1139/g77-016
2. PCR markers for Triticum speltoides leaf rust resistance gene Lr51 and their use to develop isogenic hard red spring wheat lines. Helguera M, Vanzetti L, Soria M, Khan IA, Kolmer J, Dubcovsky J. In: Crop Science, 2005 , 45:728-734. DOI: 10.2135/cropsci2005.0728
3. Catalogue of Gene Symbols for Wheat. McIntosh RA, Yamazaki Y, Devos KM, Dubcovsky J, Rogers WJ, Appels R. In: Proceedings of the 10th International Wheat Genetics Symposium. Volume 4. Instituto Sperimentale per la Cerealicoltura, Rome, Paestum, Italy.2003
