Landrace PI 374670 showed resistance to several races of the Ug99 group of Puccinia graminis f. sp tritici in a field stem rust screening nursery in Njoro, Kenya. In these tests PI 374670 showed adult infection responses between resistant (R) and moderately resistant (MR) with a low disease severity (1). In seedling tests showed infection types ranging from ‘;’ to ‘;13’ to races TTKSK, MCCFC and TPMKC. PI 374670 was collected in Bosnia and Herzegovina in 1971.
This landrace does not have any of the markers linked to the six genes associated with breeding activity (the 1RS translocation, Sr24, Sr36, Sr2, Rht-B1d, and Rht-D1b) indicating that it is most likely a real landrace and not the product of modern breeding (1).
To characterize and determine the location of this resistance locus, Babiker et al. (2) developed 216 BC1F2 families, 192 double haploid (DH) lines, and 185 recombinant inbred lines (RILs) by crossing PI 374670 and the susceptible line LMPG-6. The DH lines were tested in field stem rust nurseries in Kenya and Ethiopia and genotyped with the 90K wheat iSelect SNP genotyping platform (81,587 SNP markers, Illumina).
The seedling resistance locus of PI 374670 mapped to chromosome arm 7AL where a major QTL for field resistance was found in a 7.7 cM interval. This QTL (designated as QSr.abr- 7AL) explained 34–54 and 29–36 percent of the variation in Kenya and Ethiopia, respectively. The mapping results were verified in the RIL population (185 lines) with a set of 11 linked and flanking SNP markers converted to KASP assays. Four of these SNPs mapped to the resistance locus and the other seven were distributed around the locus.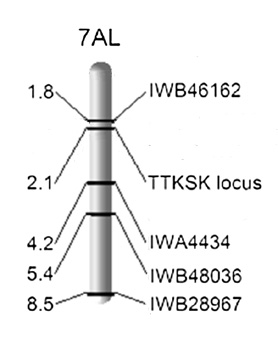
One additional QTL with four significant markers, designated QSr.abr-2DS, was detected on the short arm of chromosome 2D across a 32.5 cM interval using the Ethiopia data. This QTL explained 16–21 % of the phenotypic variation in Ethiopia.
KASP markers for QSr.abr- 7AL
In the PI 374670 x LMPG-6 population QSr.abr- 7AL was tightly linked to SNP marker IWB46162 (0.3 cM proximal) and 1.6, 3.8, and 7.7 cM proximal to SNP markers IWB19694, IWB27289, and IWB28967.
IWB28967
|
Primer name |
Primer sequence |
Allele |
Parent |
|
IWB28967_ALA |
AAACAACGGGTTCTTGCAGAGCAT |
A |
PI374670 |
|
IWB28967_ALG |
ACAACGGGTTCTTGCAGAGCAC |
G |
LMPG-6 |
|
IWB28967_C1 |
CGATAAAGGAGATATCTTCCTGCAAGTAT |
|
|
IWAB8036
|
Primer name |
Primer sequence |
Allele |
Parent |
|
IWAB8036_ALA |
ATCTCGTTTCCATTCATCTTGTACTTATA |
A |
PI374670 |
|
IWAB8036_ALG |
CTCGTTTCCATTCATCTTGTACTTATG |
G |
LMPG-6 |
|
IWAB8036_C1 |
AGCCAGTTGCTCCCACTCTATGTTT |
|
|
IWA4434
|
Primer name |
Primer sequence |
Allele |
Parent |
|
IWAB8036_ALT |
GGCAGCAAGAAGAGAAAGAAAGGATT |
T |
LMPG-6 |
|
IWAB8036_ALC |
GCAGCAAGAAGAGAAAGAAAGGATC |
C |
PI374670 |
|
IWAB8036_C1 |
CAGCGGCCTTCACCTGGGCTT |
|
|
IWB46162
|
Primer name |
Primer sequence |
Allele |
Parent |
|
IWB46162_ALT |
GAACTACGACAGCGTCTGGATCA |
T |
PI374670 |
|
IWB46162_ALC |
AACTACGACAGCGTCTGGATCG |
C |
LMPG-6 |
|
IWB46162_C1 |
ATCAACCCATGCTTTTGAAGAAGGAAATTA |
|
|
Sequences of the allele-specific primers do not include the tail sequences that interact with the fluor-labeled oligos in the KASP reaction mix. Only the SNPs that were significantly associated with the Ug99 resistance locus in the PI374670 x LMPG-6 populations are shown. There are other close SNP markers that could be useful in other crosses. For a complete list, the reader is referred to Babiker et al. (2).
For more information on KASP protocols, please visit this link.
Conditions presented here should be considered only as a starting point of the PCR optimization for individual laboratories.
References
1. Field resistance to the Ug99 race group of the stem rust pathogen in spring wheat landraces. Newcomb M, Acevedo M, Bockelman HE, Brown-Guedira G, Jackson EW, Jin Y, Njau P, Rouse MN, Singh D, Wanyera R, Goates BJ, Bonman JM. In: Plant Disease, 2013, 97:882–890. DOI:10.1094/PDIS-02-12-0200-RE.
2. Mapping resistance to the Ug99 race group of the stem rust pathogen in a spring wheat landrace. Babiker EM, Gordon TC, Chao S, Newcomb M, Rouse MN, Jin Y, Wanyera R, Acevedo M, Brown Guedira G, Williamson S, Bonman JM. In: Theoretical and Applied Genetics, 2015, published online. DOI:10.1007/s00122-015-2456-6
