The dwarfing alleles of genes that regulate plant height were essential for the success of the "Green Revolution" because they induced plants with reduced height even with a higher input of fertilizers. Many modern wheat cultivars carry semi-dominant dwarfing alleles at the Reduced height (Rht) loci on chromosome arms 4BS (Rht-B1) and 4DS (Rht-D1). These variants are known as gibberellic acid (GA) insensitive alleles because they encode repressors of the GA signaling pathway of growth promotion.
Despite its usefulness, the use of GA-insensitive variants is not recommended in environments that negatively affect seedling emergence, like dryland regions, because they tend to restrict coleoptile elongation and can also cause reductions in biomass production and yield potential. In these cases, the recommendation is to reduce plant height with GA-sensitive genes. Also, the recent identifications of different alleles with increased grain number and grain size renewed the interest in GA-sensitive genes, because the higher yield will require higher biomass to support more grains or heavier ones.
In 2018 Mo et al (1) identified a QTL (QHt.ucw-6AS) affecting plant height located in the proximal region of chromosome arm 6AS. The locus was designated Rht25 and two alleles were initially found, Rht25a (height-increasing allele) and Rht25b (dwarfing allele).
In 2023 Zhang et al. (2) determined that the gene underlying the Rht25 locus is TraesCS6A02G156600. This gene was designated it PLATZ-A1 and its function was confirmed. The authors found that PLATZ-A1 interacts physically and genetically with RHT1 with a negative interaction: the dwarfing alleles of both genes have stronger effects on plant height in the presence of the normal height of the other. They also analyzed a large panel of tetraploid and hexaploid wheats to characterize its natural variation. No dwarfing allele was found in tetraploid wheats and five in hexaploids (Rht25b, Rht25c, Rht25d, Rht25e and Rht25f) which induce significant reductions in plant height of 10 to 14% and a small negative effect in grain size:
- Rht25b: 13-bp deletion found in the third exon (CS RefSeq v1.1 6A: 145,651,904 - 145,651,916) of PLATZ-A1 in UC1110, the cultivar where the first dwarfing allele of Rht25 was found (1)
- Rht25c: 19-bp deletion and the second one is a 19-bp deletion (CS RefSeq v1.1, 6A:145,651,892 -145,651,910). It is present in McNeal
- Rht25d: splice site mutations detected in the cultivar Patwin-515HP
- Rht25e: 4-bp deletion in the third exon of PLATZ-A1 found in Chinese Spring (CS RefSeq v1.1, a CAGG deletion between 6A:145,651,864 and 145,651,865, on the - strand)
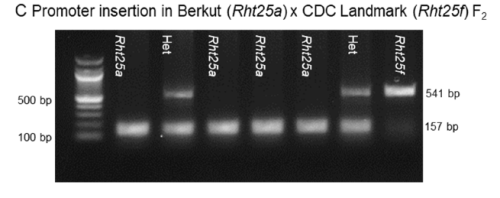
- Rht25f: 384-bp insertion in the promoter region 130 bp upstream of the start codon in CDC Landmark (CS RefSeq v1.1, 6A:145,654,767 - 145,654,768).
Markers for Rht25 variants
Zhang et al. (2) developed two PCR / PCR-CAPS and KASP markers to discriminate the natural variants of Rht25.
PCR / CAPS marker for the variants on the coding region
Primer sequences:
PlzAF2 5'-CTGTATATGCATGTGTGTCTC-3'
PlzAR1 5'-GAGGCAGCAAAAGTGGAAGG-3'
The forward primer hybridizes in the intron region.
PCR conditions
- Denaturing step: 3 min at 95°C
- 35 cycles of: [95°C 20s, 57°C 20s, 72°C 30s]
- Extension step: 5 min at 72°C
Expected Products
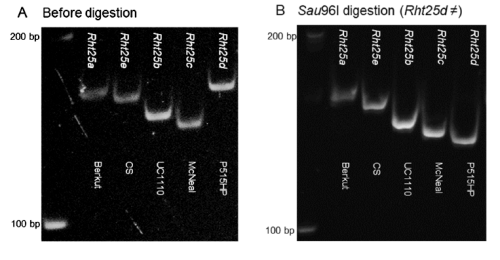
- Separate in a 6% polyacrylamide gel. Results for alleles and typical cultivars:
- Rht25a: Berkut allele, normal height. 154 bp
- Rht25b: UC110 allele, 141 bp
- Rht25c: McNeal allele, 135 bp
- Rht25d: Patwin-515HP, 154 bp (*)
- Rht25e: Chine spring, 150 bp
* To differentiate Rht25a and Rht25d digest with SauI and separate in the same type of gel
PCR marker for Rht25f (insertion on the promoter region)
Primer sequences:
platz6A-CDC-F1 5'-CACATCTACATACATCCCCATCC-3'
platz6A-CDC-F1 5'-GCAATTCTCCTCTTGGTTGGTC-3'
PCR conditions
- Denaturing step: 3 min at 95°C
- 35 cycles of: [95°C 20s, 58°C 20s, 72°C 40s]
- Extension step: 5 min at 72°C
Expected Products
- Separate in a 1.5% or 2% agarose gel.
- Rht25a: Berkut allele, normal height. 157 bp
- Rht25f: CDC Landmarl allele, 541 bp
KASP markers for Rht25 variants
KASP Marker for PLATZ-A1; 13-bp deletion(Rht25b)
|
Plz-13bp-wt-FAM |
5'- GAAGGTGACCAAGTTCATGCTGCCGAGGCCTCAGTCCA -3' |
|
Plz-13bp-mut-VIC |
5'- GAAGGTCGGAGTCAACGGATTGCCGAGGCCTCAGTCGG -3' |
|
Plz-del-common |
5'- TCCGACGGGACAATTAGGC -3' |
PCR conditions
- 94 °C 15 min
- 94 °C 20 s, 65 °C to 57 °C 1 min for 10 cycles
- 94 °C 20 s
- 57 °C 1 min for 35 cycles
- 12 °C forever
KASP Marker for PLATZ-A1 19-bp deletion (Rht25c)
|
Plz-19bp-wt-FAM |
5'- GAAGGTGACCAAGTTCATGCTcctcagtccaggccattc -3' |
|
Plz-19bp-mut-VIC |
5'- GAAGGTCGGAGTCAACGGATTcctcagtccaggccgtct -3' |
|
Plz-del-common |
5'- TCCGACGGGACAATTAGGC -3' |
PCR conditions
- 94°C 15 min
- 94°C 20 s, 65°C to 57°C 1 min for 10 cycles
- 94°C 20 s, 57 °C 1 min for 35 cycles
- 12°C forever
KASP Marker for PLATZ-A1 acceptor splice site mutation(Rht25d)
|
Plz-splice-wt-FAM |
5'- GAAGGTGACCAAGTTCATGCTactgtatatgcatgtgtgtctcA -3 |
|
Plz-splice-mut-VIC |
5'- GAAGGTCGGAGTCAACGGATTactgtatatgcatgtgtgtctcG -3' |
|
Plz-splice-common |
5'- CTTGAATGGCCTGGACTGAG -3' |
PCR conditions
- 94°C 15 min
- 94°C 20 s, 65°C to 55°C 1 min for 10 cycles
- 94°C 20 s 57°C 1 min for 35 cycles
- 12°C forever
Dominant marker for the mutant allele
KASP Marker for PLATZ-A1 4-bp deletion (Rht25e)
|
Plz-4bp-wt-FAM |
5'- GAAGGTGACCAAGTTCATGCTACATGTGACAGGATCCTCcag -3' |
|
Plz-4bp-mut-VIC |
5'- GAAGGTCGGAGTCAACGGATTACATGTGACAGGATCCTCAGC -3' |
|
Plz-del-common |
5'- TCCGACGGGACAATTAGGC -3' |
PCR conditions
- 94°C 15 min
- 94°C 20 s, 65°C to 57°C 1 min for 10 cycles
- 94°C 20 s, 57°C 1 min for 35 cycles
- 12°C forever
KASP marker of PLATZ-A1 promoter (Rht25f)
|
plz-pro-ins-FAM |
5'- GAAGGTGACCAAGTTCATGCTccatgcacgcgaaagatcaa -3' |
|
plz-pro-wt-VIC |
5'- GAAGGTCGGAGTCAACGGATTgaaaacaaaagagagatcaaacc -3' |
|
plz-pro-common |
5'- GCAATTCTCCTCTTGGTTGGT -3' |
PCR conditions
- 94°C 15 min
- 94°C 20 s, 68°C to 60°C 1 min for 10 cycles
- 94°C 20 s, 60 °C 1 min for 35 cycles
- 12°C forever
References
1. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Mo Y, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J. In: Theoretical and Applied Genetics, 2018, 131, 2021-2035. DOI:10.1007/s00122-018-3130-6.
2. Wheat plant height locus RHT25 encodes a PLATZ transcription factor that interacts with DELLA (RHT1). Zhang J, Li C, Zhang W, Zhang X, Mo Y, Tranquilli GE, Vanzetti LS, Dubcovsky J. In:
bioRxiv 2023.01.05.522836. DOI: 0.1101/2023.01.05.522836
