Contributed by W. Zhang, J. Dubcovsky
Markers for Sr13
dCAPS marker
Sr13 is a stem rust resistance gene present in several Triticum turgidum ssp. durum cultivars. Its main sources are the Ethiopian land race ST464 and the T. turgidum ssp. dicoccon L. (emmer wheat) germplasm Khapli (1,2). In tetraploid wheat cultivars, Sr13 is located on the long arm of chromosome 6A (3). Sr13 is the only known gene effective against the TTKS complex of Puccinia graminissp. tritici: the TTKSK (Ug99) race and its variants, TTKST and TTTSK. Currently this gene is the only one effective against the TTKS complex within the US durum wheat adapted cultivars, it is present in some common durum cultivars like Kofa, Kronos, Langdon, Medora and Sceptre. Sr13 is especially frequent in germplasm originated in durum breeding programs from North Dakota. This gene confers a moderate type of resistance, so it is recommended to deploy this gene pyramided with other resistance genes, which will also extend the durability of the individual genes. Despite being a frequent gene in durum lines, Sr13 has had a more restricted use in common wheat lines in the US, in contrast to its more extensive use in Australian hexaploid germplasm.
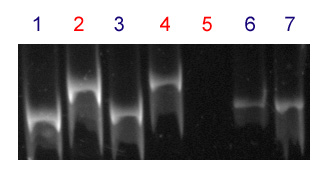
Zhang and Dubcovsky (personal communication, 4) developed a perfect marker for Sr13. They designed a derived cleaved amplified polymorphic sequence (dCAPS) marker that yields a 244-bp band in accessions carrying the resistant haplotypes and a 265-bp band or no amplificaton in accessions carrying the susceptible haplotypes.
Primers sequences:
Sr13-F 5'- TTC TTG GCT CAG AAG ACA CAT G -3'
Sr13-R 5'- AAG TCA TCA TCA TCA TTC CCG C -3'
Annealing temperature: 58°C.
After amplification, digest with restriction enzyme Hha I
Expected products
After digestion of the amplification product with restriction enzyme Hha I, accessions carrying the resistant haplotype yield a 244-bp band; and a 265-bp band in accessions carrying the susceptible haplotypes. Lack of PCR amplification with these primers is also indicative of a susceptible allele (Figure 1).
KASP markers
An alternative set of three markers based on KASP technology was developed at CIMMYT (Susanne Dreisigacker, personal communication).
These markers require two different PCR protocols: CIM_PCR_WMB002 and CIM_PCR_WMB003, which are shown below the marker information.
Sr13R/S_SNP
|
FAM primer |
agtcatcatcatcattcccccG |
|
VIC primer |
agtcatcatcatcattcccccA |
|
Common primer reverse |
gcccaggaagacaaggaagt |
|
PCR protocol |
CIM_PCR_WMB003 |
|
Favorable allele |
G |
|
CONTROL |
G: KRONOS A: UC1113 |
Sr13a/b_SNP
|
FAM primer |
ggaatgtatatgtcatgtccgacG |
|
VIC primer |
ggaatgtatatgtcatgtccgacT |
|
Common primer reverse |
cctctgcgtacgtgattacct |
|
PCR protocol |
CIM_PCR_WMB002 |
|
Favorable allele |
T |
|
CONTROL |
G: UC1113 T: KOFA |
Sr13a/c_SNP
|
FAM primer |
atccatccTatctccccaCctA |
|
VIC primer |
atccatccTatctccccaCctC |
|
Common primer reverse |
tccgatgcCtgaggaagtttaaA |
|
PCR protocol |
CIM_PCR_WMB002 |
|
Favorable allele |
A |
|
CONTROL |
A: ALTAR84 C: UC1113 |
PCR protocol CIM_PCR_WMB002
- Denaturing step: 95°C, 15 min
- Touchdown step (10 cycles):
- 95°C, 20 sec
- 65°C, 25 sec (drop 1°C each cycle)
- Amplification step (30 cycles)
- 95°C, 10 sec
- 57°C, 60 sec
PCR protocol CIM_PCR_WMB003
- Denaturing step: 94°C, 15 min
- Touchdown step (10 cycles):
- 94°C, 20 sec
- 61°C, 25 sec (drop 0.6°C each cycle)
- Amplification step (30 cycles)
- 94°C, 10 sec
- 55°C, 60 sec
- 72°C, 10 sec
Legacy markers
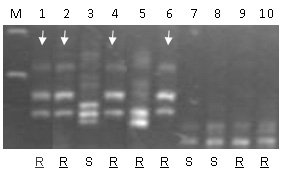
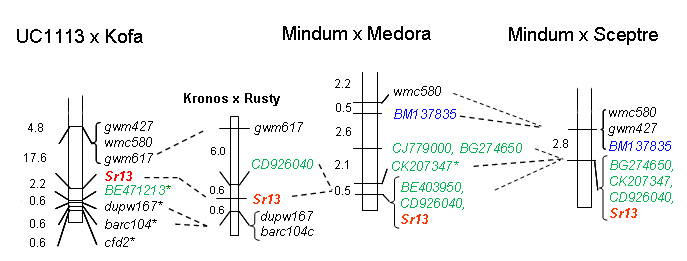
Simons et al. (3) constructed four mapping populations to precisely map Sr13 and to find molecular markers useful for breeding. These authors found that Sr13 is closely linked to several polymorphic molecular markers but they have different alleles for each donor, suggesting that the breeding programs historically used different sources of Sr13 or that independent recombination events occurred between loci. Taking together the data from the four mapping populations, the closer common markers are Xgwm427 and Xwmc580. However, considering each population separately, it is possible to find better markers. For example EST-derived marker BE403950 was completely linked to Sr13 in a Mindum x Medora population.
Table 1 below contains the information for several PCR markers (SSR and EST-derived) that map close to Sr13. The band sizes of the marker alleles vary with the donor parent considered. Also the alleles in the non-Sr13 susceptible lines are variable. Note that the current markers are not diagnostic in all the genetic backgrounds and, therefore, they cannot be used to predict the presence of Sr13in an unknown set of germplasm. These markers can be used to follow the Sr13 resistant alleles in segregating populations including some of the parental lines with any of the known Sr13 sources indicated above. The alleles associated with Sr13 from each marker and each genetic background is summarized in Table 2. Users are advised to carefully check the alleles present in the Sr13 and non-Sr13parents of their breeding lines before routine application.
| Marker | Type | Primer sequences | Notes |
| Xbarc104b | SSR | BARC104b-F GCGCTTCCAAGGCTTAGAGGCT BARC104b-R GGACCAGGCATGTCTACCCT |
Annealing temp.: 50°C |
| Xbarc104c | SSR | BARC104c-F GCATGTTTCCCATCCCTTTA BARC104c-R GCCTTCCTCCCTTTTGAAAC |
Annealing temp.: 50°C |
| Xwmc580 | SSR | WMC580-F AAGGCGCACAACACAATGAC WMC580-R GGTCTTTTGTGCAGTGAACTGAAG |
Annealing temp.: 60°C |
| Xdupw167 | SSR | dupw167-F CGGAGCAAGGACGATAGG dupw167-R CACCACACCAATCAGGAACC |
Annealing temp.: 54°C |
| XCK207347 | EST | CK207347-F TTACGGGCCACAAACAATCT CK207347-R AGCTCTCATCCATCCAGGAA |
Touch down from 60°C to 55°C |
| XCD926040 | EST | CD926040-F GTTGGCTTGGCTACTGCTTT CD926040-R AGCATTCAGCTCTGTGAGCA |
Touch down from 60°C to 55°C |
| XBE403950 | EST | BE403950-F GGAACATGTTGACGCTGTTG BE403950-R AACACTGTTCCCGAAGTTGG |
Touch down from 60°C to 55°C |
Table 1. Primers and PCR conditions for markers associated with Sr13
|
Accession |
Origin |
wmc580 |
CK207347 |
CD926040 |
BE403950 |
dupw167 |
TTKSK |
|
Sr13 present |
|
|
|
|
|
|
|
|
Khapli (CItr4013) |
NSGCa |
293 |
1000 |
851 |
691 |
230 |
;2- |
|
W2691Sr13 |
NSGCa |
326 |
1000 |
851 |
691 |
245 |
2 |
|
Langdon |
ND |
293 |
1000 |
- |
691 |
249 |
2 |
|
Kofa |
WestBred |
293 |
1000 |
855 |
723 |
230 |
2 |
|
Kronos |
AZ Pl. Breed. |
293 |
1135 |
855 |
- |
245 |
2+ |
|
Medora |
Canada |
317 |
1000 |
855 |
723 |
230 |
2 |
|
Sceptre |
Canada |
317 |
1000 |
855 |
723 |
230 |
2 |
|
ST464-C1 |
ND |
293 |
1000 |
- |
691 |
249 |
2+ |
|
Sr13 absent |
|
|
|
|
|
|
|
|
Rusty |
ND |
293 |
993 |
845 |
- |
230 |
4 |
|
Mindum |
MN |
335 |
1135 |
851 |
691 |
230 |
4 |
|
UC1113 |
CA |
317 |
1000 |
855 |
723 |
249 |
4 |
Table 2 Allele sizes for different markers in different germplasm known to carry Sr13. The markers are not diagnostic and should be used in populations with known resistance sources.
a: NSGC – USDA-ARS, National Small Grains Collection, Aberdeen, ID.
References
1. The inheritance of rust resistance: IX. The inheritance of resistance to races 15B and 56 of stem rust in the wheat variety Khapstein. Knott DR. In: Canadian Journal of Plant Sciences, 1962, 42:415-419.
2. Chromosomal locations of genes for stem rust resistance in monogenic lines derived from tetraploid wheat accession ST464. Klindworth DL, Miller JD, Jin Y, Xu SS. In: Crop Science, 2007, 47:1441-1450. DOI:10.2135/cropsci2006.05.0345.
3. Genetic mapping of stem rust resistance gene Sr13 in tetraploid wheat (Triticum turgidum ssp. durum L.). Simons K, Abate Z, Chao S, Zhang W, Rouse M, Jin Y, Elias E, Dubcovsky J. In: TAG Theoretical and Applied Genetics, 2011, 122:649-658. DOI: 10.1007/s00122-010-1444-0.
4. Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Zhang W, Chen S, Abate Z, Nirmala J, Rouse MN, Dubcovsky J. In: Proceedings of the National Academy of Sciences. In press. DOI: 10.1073/pnas.1706277114.
