Contributed by Gina Brown-Guedira, Eric Olson, Mark Sorrells and and Long-Xi Yu
Markers for Sr22
The Sr22 gene was originally identified in the diploid wheat species Triticum monococcum ssp. boeoticum accession G-21 (1) and T. monococcum L. accession RL5244 (2). It was then transferred to tetraploid and hexaploid wheat through interspecific hybridizations. Sr22 is effective against Ug99. However, a yield penalty and a delay in heading date associated with the T. monococcum ssp. boeoticum chromosome segment carrying Sr22 has limited its use (3, 8). Lines with Sr22 and reduced T. monococcum segments have been recently produced (4).
Sr22 was previously mapped on the long arm of chromosome 7A (5). Three linked markers, CFA2019, CFA2123 and BARC121 (6), were used for haplotyping this locus by Yu et al. (7), CFA2123 was not always polymorphic betwenn T. monococcum and Triticum aestivum. Olson et al. (4) produced a new set of lines with reduced alien fragments and found that the closest markers flanking Sr22 in these lines are WMC633 and CFA2123.
Primers sequences:
WMC633
WMC633-F 5'- ACA CCA GCG GGG ATA TTT GTT AC -3’
WMC633-R 5'- GTG CAC AAG ACA TGA GGT GGA TT -3’
CFA2123
CFA2123-F 5'- CGG TCT TTG TTT GCT CTA AAC C -3'
CFA2123-R 5'- ACC GGC CAT CTA TGA TGA AG -3'
CFA2019
CFA2019-F 5'- GAC GAG CTA ACT GCA GAC CC -3’
CFA2019-R 5'- CTC AAT CCT GAT GCG GAG AT -3’
BARC121
BARC121-F 5'- ACT GAT CAG CAA TGT CAA CTG AA -3’
BARC121-R 5'- CCG GTG TCT TTC CTA ACG CTA TG -3’
PCR conditions for CFA2019:
- Denaturing step: 95°C, 3 min
- Amplification step (35 cycles):
- 94°C, 45 sec
- 60°C, 45 sec
- 72°C, 60 sec
- Extension step: 72°C, 4 min
PCR conditions for WMC633:
- Denaturing step: 95°C, 2 min
- Amplification step (35 cycles):
- 94°C, 30 sec
- 56°C, 30 sec
- 72°C, 45 sec
- Extension step: 72°C, 5 min
PCR conditions for CFA2123:
- Denaturing step: 95°C, 3 min
- Touchdown cycles (decrease 1°C/cycle for 15 cycles):
- 95°C, 45 sec
- 65-51°C, 45 sec
- 72°C, 60 sec
- Amplification cycles (25 cycles)
- 95°C, 45 sec
- 50°C, 45 sec
- 72°C, 60 sec
- Extension step: 72°C, 4 min
PCR conditions for BARC121:
- Denaturing step: 94°C, 3 min
- Amplification step (35 cycles):
- 94°C, 45 sec
- 50°C, 45 sec
- 72°C, 60 sec
- Extension step: 72°C, 4 min
Concentrations of the reagents used in the PCR reaction:
For a 12 µl PCR reaction:
- 2 µl DNA (20 ng/µl)
- 1.2 µl 10X Taq buffer with 1.5 mM MgCl2
- 0.96 µl of a 100 µM mixture of dNTPs
- 0.4 µl of a 10 µM mix of forward and reverse primers
- 0.18 µl Taq polymerase (0.9 U)
- 7.26 µl dH2O
Expected products:
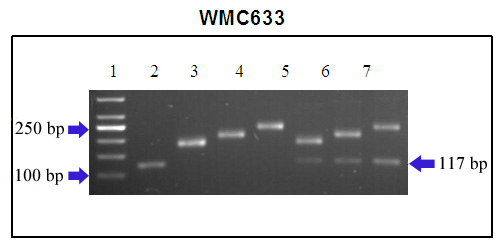
Olson et al. (4) recently mapped Sr22 4 cM distal to CFA2123 and 1-2 cM distal to WMC633. Marker WMC633 produced a 117-bp allele in the source line Sr22Tb. This allele has not been observed in any T. aestivum samples evaluated thus far (Brown-Guedira, unpublished). Several alleles are amplified by WMC633 in wheat, ranging in size from 170 bp to 260 bp. Olson et al. (4) produced an Sr22 carrying line (U5616-20-154) where recombination occurred between the resistance gene and all the SSR markers mapped in the region. In the line U5616-20-154, with the smallest T. monococcum segment, SSR marker WMC633 shows a 229-bp fragment from the hard winter wheat parent Lakin that can be used as a marker to introgress Sr22 from this line.
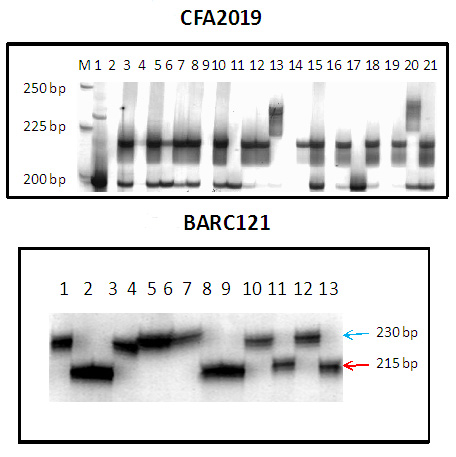
PCR amplification by CFA2019 and CFA2123 showed 238 and 234 bp fragments, respectively, in the Sr22 source germplasm, ‘Sr22TB’. BARC121 amplified two polymorphic bands of 217 and 234 bp in most germplasm. However, only the 195-bp fragment was amplified in ‘Sr22TB’. Therefore, the haplotype for Sr22 was “238-234-195” for markers CFA2019, CFA2123 and BARC121, respectively (4). Since these markers are not completely diagnostic and may produce false positive result, care must be taken when interpreting the results. Also, the allele sizes reported in Ref. 7 are slightly different.
Distribution of Donor Germplasm
Seeds of Sr22 carrying lines having a small introgression from the donor parent may be requested from Gina Brown-Guedira (Gina.Brown-Guedira@ars.usda.gov) or Mike Pumphrey (M.Pumphrey@wsu.edu).
References
1. Transfer of stem rust seedling resistance from wild diploid einkorn to tetraploid durum wheat by means of a triploid hybrid bridge. Gerechter-Amitai ZK, Wahl I, Vardi A, Zohary D. In: Euphytica, 1971, 2:281–285.
2. Inheritance of stem rust resistance transferred from diploid wheat (Triticum monococcum) to tetraploid and hexaploid wheat and chromosome location of the gene involved. Kerber ER, Dyck PL. In: Canadian Journal of Genetics and Cytology, 1973, 15:397–409.
3. Grain yields of near isogenic lines with added genes for stem rust resistance. The TT, Latter BDH, McIntosh RA, Ellison FW, Brennan PS, Fisher J, Hollamby GJ, Rathjen AJ, Wilson RE. In: Miller TE, Koebner RMD (eds). Proceedings of the 7th International Wheat Genetics Symposium, 1988, Bath Pres, Bath, UK: pp. 901-909.
4. Development of wheat lines having a small introgressed segment carrying stem rust resistance gene Sr22. Olson EL, Brown-Guedira G, Marshall D, Stack E, Bowden RL, Jin Y, Rouse M, Pumphrey MO.In: CropScience, 2010, 50:1823-1830.
DOI:10.2135/cropsci2009.11.0652.
5. Molecular mapping of stem and leaf rust resistance in wheat. Khan R, Bariana H, Dholakia B, Naik S, Lagu M, Rathjen A, Bhavani S, Gupta V. In: Theoretical and Applied Genetics, 2005, 111:846-850. DOI: 10.1007/s00122-005-0005-4.
6. Genetic mapping of a Triticum monococcum derived powdery mildew resistance gene in common wheat. Miranda LM, Perugini L, Srnic G, Brown-Guedira G, Marshall D, Leath S, Murphy JP. In: Crop Science, 2007, 47:2323-2329. DOI: 10.2135/cropsci2007.01.0053.
7. Haplotype diversity of stem rust resistance loci in uncharacterized wheat lines Yu L-X, S. Liu, J. A. Anderson, R. P Singh, Y. Jin, J. Dubcovsky, G. Brown-Guidera, S. Bhavani, A. Morgounov, Z. He, J. Huerta-Espino, M. E. Sorrells.In: Molecular Breeding, 2010, 26:667-680. DOI:10.1007/s11032-010-9403-7.
