Contributed by T.J. Tsilo, Y. Jin and J.A. Anderson
The stem rust resistance gene Sr36 derived from Triticum timopheevi confers resistance against the Ug99 race of Puccinia graminissp. tritici. However, other races have been reported to be virulent against this gene, so it advisable to deploy this gene pyramided with other resistance genes.
Two hard red spring cultivars, W1656 (CItr 12632) and W1657 (CItr 12633), were the original sources from which Sr36 was transferred to many other cultivars worldwide. Among the genes known to provide resistance against Ug99, Sr36 is the more frequent in US breeding lines. It is located on chromosome arm 2BS, and Bariana et al. (6) described ten molecular markers linked to it. Eight of these were AFLP or RFLP markers, that are either difficult to reproduce or hard to use. The other two were microsatellite markers, Xstm773 and Xgwm271. The former yields a better amplification than the latter, but the products of amplification are difficult to score, making the differentiation between homozygous and heterozygous difficult. In addition, a KASP marker is also available for this gene (see experimental details at the bottom of this section).
PCR Marker for Sr36
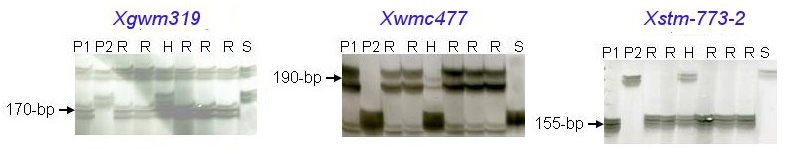
Tsilo et al. mapped three markers, Xstm773-2 (a Xstm773 derivative), Xwmc477 and Xgwm319 that were linked to Sr36, These co-dominant markers are easier to score and were validated in 76 cultivars from 12 countries (1). The authors used two mapping populations to map these markers in relation to the Sr36 locus: LMPG-6 x Sr36/9*LMPG and 'Chinese Spring' x W2691Sr36-1. In both cases a distorted segregation that favored transmission of the Sr36-carrying segment was observed. In both populations, the two markers Xstm773-2 and Xwmc477 were completely linked to Sr36. The marker Xgwm319 was completely linked to Sr36 in the LMPG-6 x Sr36/9*LMPG population and was 0.9 cM away in the 'Chinese Spring' x W2691Sr36-1 population
Primers sequences:
Xgwm319
WMS319F 5'- GGT TGC TGT ACA AGT GTT CAC G -3'
WMS319R 5'- CGG GTG CTG TGT GTA ATG AC -3'
Xwmc477
WMC477F 5'- CGT CGA AAA CCG TAC ACT CTC C -3'
WMC477R 5'- GCG AAA CAG AAT AGC CCT GAT G -3'
Xstm773-2
STM773-2F 5'- ATG GTT TGT TGT GTT GTG TGT AGG -3'
STM773-2R 5'- AAA CGC CCC AAC CAC CTC TCT C -3'
The STM773-2 primers were developed by M. Hayden & P. Sharp, who kindly provided the sequences and made them available for breeding (2).
Final concentrations of the reagents used in the PCR amplification
PCR reactions were performed in 96-well plates with 10 ul of a final reaction mixture containing
- 4.5 ng/µl genomic DNA
- 0.1 µM each primer
- 0.2 mM each dNTP
- 0.025U/µl Taq DNA polymerase (Applied Biosystems)
- 1x PCR buffer
- 1.5 mM MgCl2
Total volume: 10 µl
PCR conditions for WMS319 and WMC477 :
- Denaturing step: 94°C, 10 min
- Amplification step (35 cycles):
- 94°C, 1 min
- 55°C for WMS319, or 61°C for WMC477, 1 min
- 72°C, 2 min
- Extension step: 72°C, 10 min
PCR conditions for STM773-2 :
- Denaturing step: 94°C, 10 min
- Touchdown step 1 (7 cycles):
- 92°C, 1 min
- 64°C, 1 min
- 72°C, 1 min
- Touchdown step 2 (5 cycles)
- 92°C, 1 min
- 57°C, 1 min
- 72°C, 1 min
- Amplification step (10-25 cycles):
- 92°C, 30 s
- 55°C, 1 min
- 72°C, 1 min
- Extension step: 72°C, 10 min
Expected products:
The PCR products can be separated using polyacrylamide gels (6%, acrylamide/bisacrylamide 20:1, 8M urea, TBE pH 8.3). Microsatellite alleles linked with the presence of Sr36 have typical fragments of 170-bp (Xgwm319), 190-bp (Xwmc477) and 155-bp (Xstm773-2). For any system (Ag-stained gel or ABI), it is important to run the PCR reaction for WMC477 at an annealing temperature of 61°C.
KASP marker for Sr36
SNP ID wMAS000015
Gene Sr36
Evidence association
Primer Allele FAM ACCCAGCCGCTTGAGGCG
Primer Allele VIC CACCCAGCCGCTTGAGGCT
Primer Common CAGCGTAGTGCGCGCGGCTT
FAM allele G
VIC allele T
FAM phenotype susceptible
VIC phenotype resistant
For more information on KASP protocols, please check visit this link.
References
1. Diagnostic microsatellite markers for the detection of stem rust resistance gene Sr36 in diverse genetic backgrounds of wheat. Tsilo TJ, Jin Y, Anderson JA. In: Crop Science, 2008, 48:253-261. DOI:10.2135/cropsci2007.04.0204
2. Sequence-tagged microsatellite profiling (STMP): a rapid technique for developing SSR markers. Hayden MJ, Sharp PJ. In: Nucleic Acids Research, 2001, 29: e43. DOI:10.1093/nar/29.8.e43
