Contributed by Rohit Mago and Ian Dundas
Markers for Sr39-Lr35
The stem rust resistance gene Sr39 provides resistance to all currently known pathotypes of Puccinia graminis f. sp. tritici (Pgt) including Ug99 (TTKSK) and its variants TTKST and TTTSK, which are virulent against Sr24 and Sr36 respectively, two frequently deployed resistance genes.
Sr39 was transferred to the hexaploid wheat cultivar Marquis from Aegilops speltoides (1). The gene is located on a translocated segment of A. speltoides chromosome 2S to wheat chromosome 2B (translocation stock 2S#2). The translocated segment carries the seedling stem rust resistance gene Sr39 and an adult-plant hypersensitive leaf rust resistance gene Lr35. No current Australian wheat cultivar carries Sr39, presumably because of negative agronomic effects of this large translocation. Labuschagne et al. (2) reported that a South African wheat genotype (Karee*6/RL6082) with Lr35 (and also Sr39) showed significant increase in flour water absorption compared to the recurrent parent Karee. Translocation lines with reduced T. speltoides segments were developed that carry Sr39 and Lr35 resistance (recombinant # 247)
A combination of dominant PCR markers in repulsion phase provides a diagnostic co-dominant marker for Sr39: Sr39#22r, a dominant marker for the presence of Sr39 (3); and EST-derived marker BE500705, or alternatively Sr39#50s, two dominant markers for the absence of Sr39 (2). Markers BE500705 or Sr39#50s in combination with Sr39#22r provide a codominant marker useful to transfer the shortened A. speltoides translocation carrying Sr39 into commercial wheat cultivars and to combine this gene with others effective against TTKSK and its variants. The susceptible markers are not perfect, we found 10% linkage with Sr39#50s and one susceptible wheat variety that did not amplify with BE500705. So there is possibly some variability present in wheat.
You can find additional information on the Lr35 page
Primer sequences:
Sr39#22r
Sr39#22r-F 5'- AGA GAA GAT AAG CAG TAA ACA TG -3'
Sr39#22r-R 5'- TGC TGT CAT GAG AGG AAC TCT G -3'
BE500705
BE500705-F 5'- ATC TGT GGC AGT GTG CTC CT -3'
BE500705-R 5'- TCC TGC AAA TGC TTG TCG TT -3'
Sr39#50s
Sr39#50s-F 5'- CCA ATG AGG AGA TCA AAA CAA CC -3'
Sr39#50s-R 5'- CTA GCA AGG ACC AAG CAA TCT TG -3'
PCR conditions for Sr39#22r:
- Denaturing step: 94°C, 5 min
- Amplification step (30 cycles):
- 92°C, 30 sec
- 58°C, 30 sec
- 72°C, 40 sec
- Extension step: 72°C, 10 min
PCR conditions for BE500705:
- Denaturing step: 94°C, 3 min
- Amplification step (30 cycles):
- 92°C, 30 sec
- 56°C, 30 sec
- 72°C, 40 sec
- Extension step: 72°C, 10 min
PCR conditions for Sr39#50s:
- Denaturing step: 94°C, 3 min
- Touchdown step 1 (7 cycles, 1ºC down each cycle):
- 92°C, 30 sec
- 65°C, 30 sec
- 72°C, 40 sec
- Amplification step (30 cycles):
- 94°C, 30 sec
- 58°C, 30 sec
- 72°C, 40 sec
- Extension step: 72°C, 10 min
Final concentrations of the reagents used in the PCR amplification
For a 20-µl PCR reaction:
- 2 µl DNA (50-100 ng/µl)
- 2 µl 10X Taq buffer (with MgCl2 )
- 0.4 µl dNTPmix (10mM each)
- 2 µl forward primer (5 µM)
- 2 µl reverse primer (5 µM)
- 1 µl Taq
- dH2O to 20 µl
Expected products:
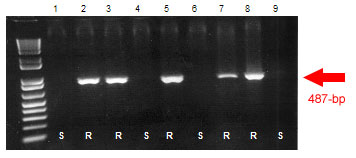
Sr39#22r: STS Sr39#22r is a dominant marker that amplifies a single 487-bp band linked to the A. speltoides translocation containing Sr39.
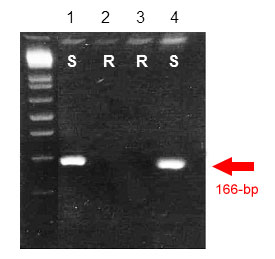
BE500705: EST BE500705 is a dominant marker that amplifies a single 166-bp band also linked to the original wheat segment (susceptible allele), although at least one susceptible wheat line did not yield any amplification product. This marker can be used in combination with Sr39#22rs to generate a codominant marker.
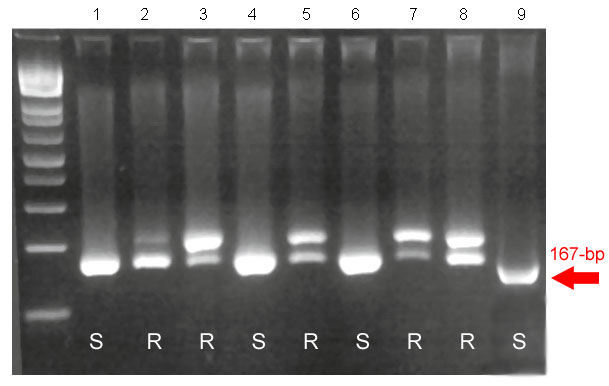
Sr39#50s: STS Sr39#50s is a dominant marker that amplifies a single 167-bp band locus in susceptible lines and two bands in homozygous resistant recombinant lines. However, only the lower band, with a size of size 167-bp, is amplified in heterozygous recombinant plants. The association between this marker and the lack of resistance is not perfect (10% linkage). STS Sr39#50s is an alternative to BE500705, and can also be used to generate a codominant marker in combination with Sr39#22rs.
Distribution of Donor Germplasm
Sr39 donor lines with reduced A. speltoides segments have been described in Mago et al. (3). To obtain seed of these recombinant donor lines for your breeding program, please contact:
Dr. Ian S. Dundas
The University of Adelaide
ian.dundas@adelaide.edu.au
References
1. Transfer to hexaploid wheat of linked genes for adult-plant leaf rust and seedling stem rust resistance from amphiploid of Aegiolops speltoides x Triticum monoccum. Kerber ER, Dyke PL. In: Genome, 1990, 33:530-537.
2. The influence of leaf rust resistance genes Lr29, Lr34, Lr35 and Lr37 on bread making quality in wheat. Labuschagne MT, Pretorius ZA, Grobbelaar B. In: Euphytica, 2002, 124: 65-70. DOI:10.1023/A:1015683216948
3. Development of wheat lines carrying stem rust resistance gene Sr39 with reduced Aegilops speltoides chromatin and simple PCR markers for marker-assisted selection. Mago R, Zhang P, Bariana HS, Verlin DC, Bansal UK, Ellis JG, Dundas IS. In:Theoretical and Applied Genetics, 2009, 124:65-70. DOI:10.1007/s00122-009-1146-7
