Contributed by Colin Hiebert and Tom Fetch
Hiebert et al. (1) reported that Canadian hard red Spring cultivars “Peace” and “AC Cadillac” are resistant to Puccinia graminis f. sp.tritici race Ug99 and its variants at the seedling stage and in the field. Analyzing two mapping populations derived from each resistant cultivar, the authors found an apparently new gene temporarily designated SrCad that maps on chromosome arm 6DS. In field trials in Kenya two populations carrying SrCad only presented high levels of resistance to stem rust when combined with the leaf rust resistance gene Lr34. In its absence, SrCad only conferred moderate resistance against to Ug99 and related races. Lr34 is a gene that confers partial but broad spectrum resistance against several pathogens and it is known to interact positively with other leaf rust resistance genes (2) and stem rust resistance genes (3)
Two other stem rust resistance genes were mapped on the short arm of chromosome 6D, Sr5 and Sr42. Although gene Sr5 is not resistant to Ug99, it cannot be discarded that SrCad is a new allele. Sr42 is resistant to Ug99 and its resistance phenotype is similar to that of SrCad. Further tests are being carried out to determine whether SrCad is a new gene or an allele of any of those previously described genes (1).
Markers for SrCad
On the distal side, the closest polymorphic marker is CFD49 at a distance of 7.5 cM. The PCR marker FSD_RSA, linked to the bunt (Tilletia tritici) resistance gene Bt10, (4) is aproximately 1.6 cM proximal from the SrCad locus and is the recommended marker for molecular breeding.
Primers sequences:
FSD_RSA
FSD 5'- GTT TTA TCT TTT TAT TTC -3'
RSA 5'- CTC CTC CCC CCA -3'
PCR conditions for FSD_RSA:
- Denaturing step: 94°C, 3 min
- Amplification step (35 cycles):
- 94°C, 30 sec
- 44°C, 1 min 45 sec
- 72°C, 2 min
- Extension step: 72°C, 10 min
Final concentrations of the reagents used in the PCR amplification
- 1X PCR buffer
- 1.5 mM MgCl2
- 0.2 mM each dNTP
- 12 pmol FSD forward primer
- 3.5 pmol RSA reverse primer
- 1U Taq DNA polymerase
- 50 ng of genomic DNA
Expected products:
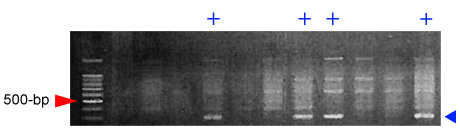
References
1. Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars 'Peace' and 'AC Cadillac'.Hiebert CW, Fetch TG, Zegeye T, Thomas JB, Somers DJ, Humphreys DG, McCallum BD, Cloutier S, Singh D, Knott DRIn: TAG Theoretical and Applied Genetics, 2010. DOI:10.1007/s00122-010-1430-6
2. Effect of the gene Lr34 in the enhancement of resistance to leaf rust of wheat. German SE, Kolmer JA. In: TAG Theoretical and Applied Genetics, 1992, 84:97-105. DOI:10.1007/BF00223987
3. Leaf rust resistance gene Lr34 associated with nonsuppression of stem rust resistance in the wheat cultivar Canthatch. Kerber ER, Aung T. In: Phytopathology, 1999, 89:518–521 DOI:10.1094/PHYTO.1999.89.6.518
4. Development of a PCR marker for rapid identification of the Bt-10 gene for common bunt resistance in wheat.. Laroche A, Demeke T, Gaudet DA, Puchalski B, Frick M, McKenzie R. In: Genome, 2000, 43:217-223. DOI:10.1139/gen-43-2-217
