The SrTA10187 gene originated from Aegilops tauschii accession TA10187, which was collected in Turkmenistan. It is resistant to races TTKSK, TTKST (both Ug99-related) TTTTF, TPMKC RKQQC and QTHJC (the last two are endemic to North America).
SrTA10187 was transferred to KS05HW14, a high yielding hard white winter wheat line resistant to soilborne wheat mosaic virus, wheat streak mosaic virus, and moderately resistant to stripe rust, but susceptible to stem rust races QTHJC, RKQQC, TPMKC, and TTKSK (1).
The gene transfer from A. tauschii to Triticum aestivum was carried out using a direct crossing technique facilitated by the existence of homologous recombination between the D genome chromosomes of both species (2). Briefly, lines of diploid A. tauschii were crossed with the adapted hexaploid wheat cultivar. Since the resulting F1 hybrids (ABDD genomic constitution) frequently show an aberrant endosperm formation, the procedure is followed by embryo rescue and then backcrossed to the adapted wheat cultivar.
The SrTA10187 locus is located on chromosome arm 6DS. A high resolution mapping population including KS05HW14, TA10187, and 1,060 BC3F2 individuals was used to map its location, which is shown in the figure below (3). Sr5, SrCad, SrTmp and Sr42 are stem rust resistance genes located close to SrTA10187 on chromosome 6D, but susceptibility tests suggest they are different loci. Further allelism tests are underway to confirm SrTA10187 is a unique locus.
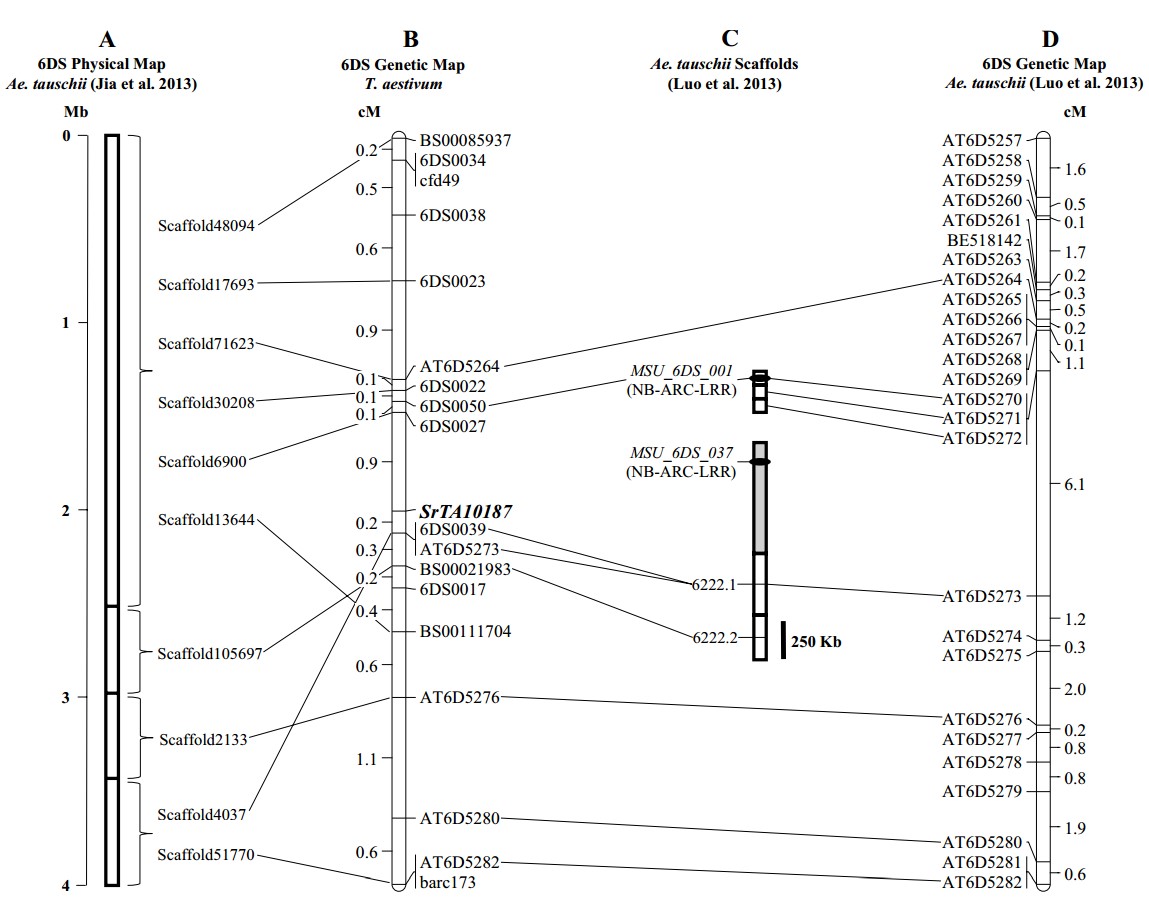
Markers for SrTA10187
The KASPTM markers 6DS0027 and 6DS0039 flank SrTA10187 at 0.9 and 0.2 cM, respectively. The STS marker, 6DS0050, is located 1.0 cM distal to SrTA10187 and can be assayed by gel electrophoresis.
Primers sequences:
KASPTM and STS markers used for fine mapping of SrTA10187 on wheat chromosome 6DS
|
Marker ID |
Marker Expression |
Primer Name |
Primer Sequence (c) |
Allele |
Parent |
|
6DS0050 (b) |
Dominant |
6DS0050_F |
AAAGGGAGTTGATATGGATCTGT |
- |
TA10187 |
|
6DS0050_R |
CAAGCTGGTTAAAATAGCTCTCA |
||||
|
6DS0027 (a) |
Dominant |
6DS0027_A1 |
AATGTCATCTCAGTAAAACAAATTGCC |
G |
TA10187 |
|
6DS0027_A2 |
AATGTCATCTCAGTAAAACAAATTGCT |
A |
KS05HW14 |
||
|
6DS0027_C |
CATATTTTGGCCTTTCTGTCAGAGAGT |
||||
|
6DS0039 (a) |
Codominant |
6DS0039_A1 |
AAACTGCAGAGCATTTCATTTTTT |
A |
TA10187 |
|
6DS0039_A2 |
AAACTGCAGAGCATTTCACTTTTG |
C |
KS05HW14 |
||
|
6DS0039_C |
GGTTACATCATAAACTACATGTGCATAG |
||||
|
AT6D5273 (b) |
Dominant |
AT6D5273_A1 |
ACTGTGAGTACATTGGGACTAGAATT |
A |
KS05HW14 |
|
AT6D5273_A2 |
ACTGTGAGTACATTGGGACTAGAATC |
G |
TA10187 |
||
|
AT6D5273_C |
AAGTCTATCCCGGCTGGATGCA |
a: Source of the SNP context sequence: genotype-by-sequencing tags identified by bulk segregant analysis
b: Source of the SNP context sequence: Sequencing the Aegilops tauschii Genome (http://aegilops.wheat.ucdavis.edu/)
c: A1 and A2 primers are listed without FAM and HEX 5' end sequences, in 5’ to 3’ orientation
PCR conditions for KASP™ markers:
- Denaturing step: 94°C, 15 min
- Amplification (10 step-down cycles):
- 94°C, 20 sec
- 61-55°C, 1 min (decreasing by 0.6°C each cycle)
- Amplification (26 cycles):
- 94°C, 20 sec
- 55°C, 1 min
- HEX and FAM fluorescence detection
KASP™ PCR amplification reagents:
- For 5 µL reactions
- 10 ng of gDNA
- 0.07 µL of primer mix (12 µM of each allele-specific forward primer and 30 µM reverse primer)
- 2.5 µL of KASPTM master mix
PCR conditions for STS marker 6DS0050:
- Denaturing step: 95°C, 4 min
- Amplification (34 cycles):
- 95°C, 45 sec
- 57°C, 45 sec
- 72°C, 30 sec
STS marker 6DS0050 PCR amplification reagents:
- For 20 µL reactions
- 120 ng of gDNA
- 0.5 µL of each primer (10 µM)
- 2 µL of 10 x reaction buffer
- 0.5 µL of dNTP (10 µM)
- 0.2 µL of Taq polymerase
Expected products
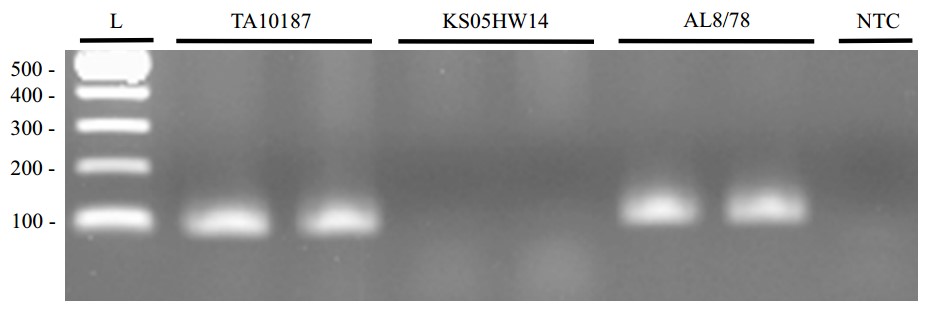
Conditions presented here should be considered only as a starting point of the PCR optimization for individual laboratories.
References
1. Introgression of stem rust resistance genes SrTA10187 and SrTA10171 from Aegilops tauschii to wheat. Olson EL, Rouse MN, Pumphrey MO, Bowden RL, Gill BS, Poland JA. In: Theoretical and Applied Genetics, 2013, 126:2477-2484. DOI: 10.1007/s00122-013-2148-z
2. Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Olson EL, Rouse MN, Pumphrey MO, Bowden RL, Gill BS, Poland JA. . In: Theoretical and Applied Genetics, 2013, 126:1179–1188. DOI: 10.1007/s00122-013-2045-5
3. Fine mapping of the stem rust resistance gene SrTA10187. Wiersma AT, Brown LK, Brisco EI, Liu TL, Childs KL, Poland JA, Sehgal SK, Olson EL. In: Theoretical and Applied Genetics, 2016, 129:2369–2378. DOI: 10.1007/s00122-016-2776-1
