Stripe rust is one of the most aggressive diseases on common wheat (Triticum aestivum L. ) and durum wheat (T. turgidum L.var. durum) worldwide. In the United States, the western states are the most affected, although is becoming more important in the central US. Stripe rust is caused by the fungus Puccinia striiformis Westend. f. sp. tritici Eriks. (P. s. tritici).
The preferred way of controlling the disease is through the use of resistant varieties. There are several genes that can express resistance to this disease. However, changes in pathogen virulence can render them useless for breeding after some time.
Yr5 was described first in 1966 by Macer in Triticum spelta album (1). This gene confers resistance to all of the races known in the US (X. M. Chen, personal communication). Yr5 is located on chromosome arm 2BL, 21 cM away from the centromere (2). Kema transferred this gene into some commercial cultivars (3), and it remains effective to a broad range of PST isolates worldwide.
Marchal et al. (7) cloned Yr5 and showed that it is allelic to YrSP and paralogous to Yr7, both of which have been overcome by multiple PST isolates. The sequence analysis revealed that Yr5 belongs to a complex resistance gene cluster on chromosome 2B encoding nucleotide-binding and leucine-rich repeat proteins (NLRs) with a distinct N-terminal zinc-finger BED domain.
Markers for Yr5
KASP marker
When comparing the Yr5 gene structure to its closer sequence-related genes, an insertion that generated an alternate STOP codon was identified (see figure below). YrSP has two SNPs and one of them generates a premature STOP codon, leading to a shorter protein. The Yr5 alleles in Claire, Cadenza and Paragon all have a similar sequence near the STOP codon.
Based on this observation a KASP assay was designed to target this insertion associated with the Yr5 allele that confers resistance (Yr5_Positive) and tested its presence in other varieties displaying a Yr5 resistance phenotype (7, also see primers and probes below). Blue and purple primers can bind all the alleles, however, because of the insertion in Yr5 they would be too far apart to allow amplification of the product in a typical KASP assays (the product is more that 700 bp). The red primer is right at the junction between the insertion and the end of the third exon, so it is specific to that structure and close enough to the purple primer to allow amplification and detection.
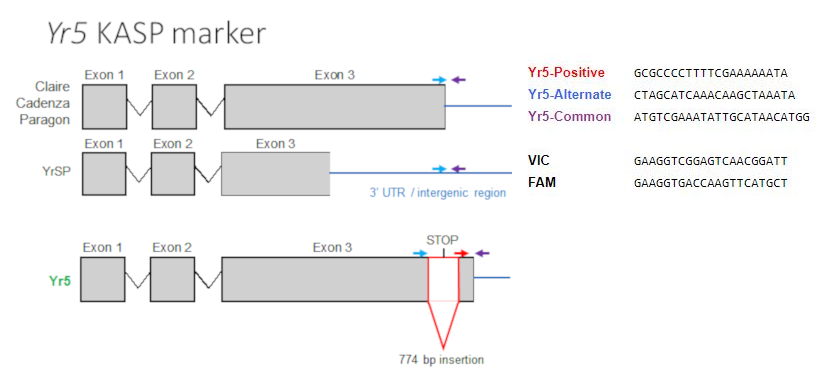
KASP probe sequences (text format)
Yr5-Positive GCGCCCCTTTTCGAAAAAATA
Yr5-Alternate CTAGCATCAAACAAGCTAAATA
Yr5-Common ATGTCGAAATATTGCATAACATGG
VIC GAAGGTCGGAGTCAACGGATT
FAM GAAGGTGACCAAGTTCATGCT
Legacy markers for Yr5
Gel based marker
The comparative analysis of Yr5 and other non-resistant aleles showed that Yr5 has a 774 bp insertion 29 bp upstream of the STOP codon. Marchal et al. (7) designed PCR primers flanking that insertion and tested them on a germplasm set to confirm the usefulness of this marker for molecular breeding.
PCR primers:
Yr5_insertion_F 5'- CTC ACG CAT TTG ACC ATA TAC AAC T -3'
Yr5_insertion_R 5'- TAT TGC ATA ACA TGG CCT CCA GT -3'
Amplification and expected products
The PCR amplification was conducted using a touchdown program: 10 cycles, -0.5 °C per cycle starting from 67°C and the remaining 25 cycles at 62°C.
Three different profiles were found: (1) a 1,281 bp product in Yr5 positive cultivars, (2) a 507 bp amplicon in the alternate Yr5allele carriers, including AvocetS-YrSP, Cadenza, and Claire, and (3) no amplification:
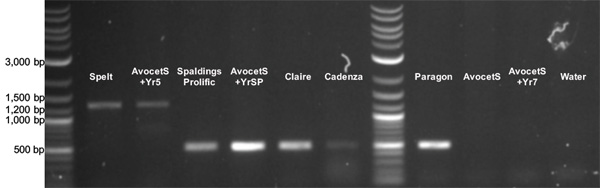
In some Yr5 lines the 1281-bp is difficult to visualize and the amplification protocol might need further optimization for specific lines and lab conditions (Xiaoqin Zhang, personal communication).
Other markers
Yan et al. found some RGAP (resistance gene analog polymorphism) markers completely linked to Yr5 (4). This type of markers are obtained by PCR amplification of sequences known to be homologous to conserved domains of genes coding for disease resistance (5). Usually, many products are obtained, and those that co-segregate with the trait in a mapping population can be used as markers. However, this type of markers are difficult to use and score, specially in a breeding environment, besides in some cases they are not polymorphic for the recurrent parent.
Later, Chen et al (6) converted two of the RGAP markers into more user friendly PCR markers. They developed two STS (sequence tagged site) and one CAPS (cleaved amplified polymorphic sequence) markers. These markers are polymorphic for a wider range of recurrent parents. The STS markers are analyzed in polyacrylamide gels, but they are easier to score than the original RGAPs, and the CAPS marker can be resolved in agarose gels.
STS markers for Yr5
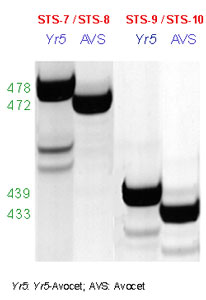
There are two STS markers available for detecting the presence of Yr5: STS-7/STS-8 and STS-9/STS-10. The PCR protocol is the same for both sets of primers
Primers:
STS-7 5'- GTA CAA TTC ACC TAG AGT -3'
STS-8 5'- GCA AGT TTT CTC CCT ATT -3'
____________________________________
STS-9 5'- AAA GAA TAC TTT AAT GAA -3'
STS-10 5'- CAA ACT TAT CAG GAT TAC -3'
PCR conditions:
- Denaturing step: 94°C, 5 min
- 45 cycles of: [94°C 30 sec, 45°C 30 sec, 72°C 45 sec]
- Extension step: 72°C 7 min
A 2.5 min ramp time is used between the 94°C denaturation and the 45°C annealing steps; for all other temperature transitions, the fastest possible ramp is used. The number of amplification cycles can be reduced to 40 if a more sensitive detection system is used.
Final concentrations of the products used in the PCR reaction:
- dNTPs: 0.2 mM each
- MgCl2: 5 mM
- primers: 0.4 µM each
- Taq DNA polymerase (Promega): 0.6 unit
- 10X PCR buffer (Promega, Mg-free): 1.5 µl
- Genomic DNA: 30 ng
Final volume: 15 µl
Expected products:
PCR products are separated in 5% denaturing polyacrylamide gel. When using the set of primers STS-7 and STS-8, Avocet produces a band of 472 bases and Yr5-Avocet a band of 478 bases. The other marker STS-9 / STS-10 yields a product of 433 for Avocet and 439 for Yr5-Avocet.
CAPS marker for Yr5
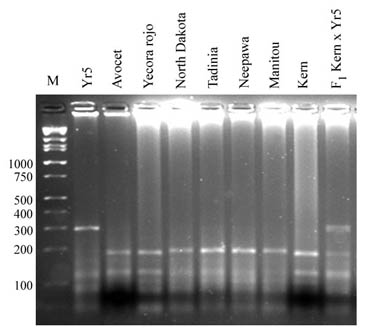
Some cultivars are not polymorphic for the STS markers described above. Besides, polyacrylamide gels are more difficult to prepare and develop than agarose gels. Chen et al. (In press) analyzed the sequence of the fragments amplified by the STS markers and found a single base pair polymorphism that generates a Dpn II restriction site in the DNA of susceptible parents. This restriction site was used to develop a CAPS marker that was tested in a large set of cultivars
Procedure:
- Run a PCR reaction with primers STS-9 and STS-10 as described
- Digest 17.5 µl of PCR product with 0.5 µl (5 U) of restriction enzyme Dpn II (New England Biolabs) and 2 µl of 10X buffer for Dpn II (New England Biolabs) for 2 h at 37°C.
- Separate products in 2.8% agarose gels.
Some laboratories reported difficulties amplifying with primer STS-9. As an alternative, the PCR amplification can be done using primers pairs STS-7/STS-8 or STS-7/STS-10. In both cases the digestion procedure is the same as indicated before, and the products are co-dominant.
Expected products:
After digestion the resistant line Yr5-Avocet yields five restriction fragments (289 bp, 63 bp, 56 bp, 20 bp and 12 bp), while Avocet and other non-Yr5 genotypes produce four restriction fragments (182 bp, 102 bp, 20 bp and 12 bp). The presence/absence pattern of the 289-bp fragment from the Yr5 line and the 182-bp fragment from the susceptible lines can be scored in agarose gels.
Additional information
Reaction to PST races of P. striiformis f. sp. tritici
| PST races | 17 | 20 | 27 | 29 | 37 | 43 | 45 | 59 | 78 |
| Avocet | S | S | S | S | S | S | S | S | S |
| Yr5 in an Avocet background | R | R | R | R | R | R | R | R | R |
References
1. The formal and monosomic genetic analysis of stripe rust (Puccinia striiformis) resistance in wheat. Macer, RCF. In: IJ. Mackey (ed.) Proc. of 2nd Int. Wheat Genet. Symp. Lund, Sweden 1963. Hereditas Suppl., 1966, 2:127-142.
2. Genetic control of yellow rust resistance in T. spelta album. Law CN. In: Plant Breeding Institute, Cambridge, Annual Report 1975, 1976, 108-109.
3. Resistance in spelt wheat to yellow rust I. Formal analysis and variation for gliadin patterns. Kema GHJ. In: Euphytica, 1992, 63:207-217.
4. Resistance gene analog polymorphism markers co-segregating with the Yr5 gene for resistance to wheat stripe rust have homology with plant disease resistance genes. Yan GP, Chen XM, Line RF, Wellings CR. In: Theoretical and Applied Genetics, 2003, 106:636-643. DOI:10.1007/s00122-002-1109-8
5. Isolation of a superfamily of candidate disease resistance genes in soybean based on a conserved nucleotide-binding site. Yu YG, Buss GR, Maroof MA. In: Proc Natl Acad Sci USA, 1996, 93:11751-11756. DOI:10.1073/pnas.93.21.11751
6. Development of Sequence Tagged Site and Cleaved Amplified Polymorphic Sequence Markers for Wheat Stripe Rust Resistance Gene Yr5. Chen XM, Soria MA, Yan GP, Sun J, Dubcovsky J. In: Crop Science, 2003, 43:2058-2064.DOI:10.2135/cropsci2003.2058
7. BED-domain containing immune 1 receptors confer diverse resistance spectra to yellow rust. Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Adamski NM, Boyd L, McIntosh R, Wulff BBH, Berry S, Lagudah E, Uauy C. In: .DOI:10.1038/s41477-018-0236-4. Alternative free link
