Achieving higher grain yields is a difficult task due to the number of genes involved, their interactions and the importance of environmental effects. This complexity determines that taken as a single trait, yield shows low heritability. Instead of improving overall yields, a more effective strategy is to focus on yield components, such as grain weight, spikes per unit area, grain number per spikelet and spikelet number per spike (SNS). Among these, SNS is an attractive trait for wheat breeders because it shows a higher heritability compared to the other yield components. For example, in one study the average heritability (H2) for SNS was 0.84 (1). Furthermore, SNS is a trait determined at an early developmental stage and, therefore, is less influenced by the environmental conditions after the terminal spikelet is formed.
In a genome-wide association study (GWAS) including 262 hexaploid spring wheat accessions, Zhang et al (1) found a highly significant association between SNS and SNP marker IWA5912 (T3/Wheat, GrainGenes) on chromosome arm 7AL.
Other GWAS and analyses of biparental populations found evidence of loci associated with SNS and grain number per spike in that region of chromosome arm 7AL in tetraploid and hexaploid wheat (1-9).
Taken together the previous studies defined a region of yield component QTLs between 672.0 and 674.3 Mb in the 7A pseudomolecule of the Chinese Spring genome released by the International Wheat Genome Sequencing Consortium (IWGSC, annotation RefSeq v1.0). Later, Kuzay et al. (10) mapped the SNS locus to a narrow region of 87-kb containing only two complete and two partial genes. Only one of these genes, TraesCS7A02G481600, had a non-synonymous polymorphism that co-segregates with differences in SNS. Besides, this gene is orthologous to the rice gene ABERRANT PANICLE ORGANIZATION 1 (APO1), which affects panicle development and spikelet number in rice (11). Hence, the wheat gene for SNS was named WHEAT ORTHOLOG of APO1, WAPO1, and was proposed as the most likely candidate gene responsible for the 7AL SNS QTL. Later, Kuzay et. al (11) using loss of function mutants and transgenic lines could functionally validate that WAPO1 is indeed responsible for the SNS phenotype.
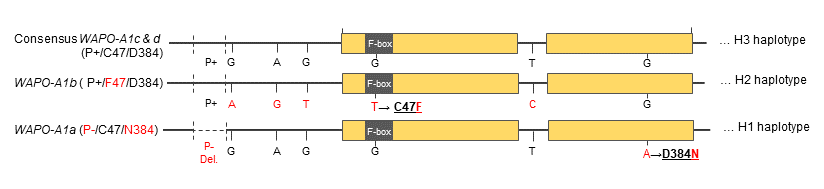
Haplotype analysis of the candidate region using available exome capture data revealed the existence of three haplotype blocks in the region: H1, H2 and H3, with H2 always associated with higher SNS than H1 and H3. Haplotype 3 was at very low frequency in both durum and common wheat, H1 was predominant in durum wheat and H2 was at high frequency in modern common wheat varieties. The increased in spikelet number in H2 was associated with increases in grain yield when the allele was introgressed in high-biomass productive varieties and those varieties were grown in environments with adequate water and nitrogen (1).
The high SNS WAPO-A1 allele from the H2 haplotype was designated as WAPO-A1b and the one present in the H1 haplotype as WAPO-A1a. Haplotype H3 included two WAPO-A1alleles, WAPO-A1c and WAPO-A1d. The protein coded by WAPO-A1b has an amino acid change, C47F, which differentiates it from the variants found in the other haplotypes. This variant is located within a conserved F-box. In addition, WAPO-A1b differed from the other by a SNP in its intron and three SNPS in the promoter region.
As a tool to facilitate the introgresson of the H2 haplotype in durum what, Kuzay et. al (11) deposited a Kronos NIL with the H2 introgression in the National Small Grains Collection with accession number PI 698810.
Markers for WAPO-A1
Kuzay et al. (10) developed two markers for allele identification. The first is a PCR marker that targets a unique 115-bp deletion in the promoter region of WAPO-A1a. The other is a dCAPS marker for the C47F polymorphism that is unique to WAPO-A1a. Using this pair of markers is it possible to differentiate WAPO-A1a and WAPO-A1b from the other alleles.
PCR marker for the 115-bp deletion
Primer sequences:
WAPO1_pro_F 5'- ACGGTTCCTCTTCCTGCTCAT -3'
WAPO1_pro_R 5'- CGGAGGCGAGGACGAGT -3'
PCR conditions
- Denaturing step: 5 min at 94°C
- 35 cycles of: [94°C 30s, 60°C 30s, 72°C 30s]
- Extension step: 7 min at 72°C
Expected Products
- Separate in a 2% agarose gel:
- WAPO-A1a: one band of approx. 100-bp
- WAPO-A1b, WAPO-A1c, WAPO-A1d : one band of approx. 200-bp
dCAPS for C47F polymorphism
Primer sequences:
WAPO1_C47F_F 5'- agctcactcactctcActccA -3'
WAPO1_C47F_R 5'- GAAGGTCGGAGTCAACGGATTgaggaaggacggcgtcgggatg -3'
PCR conditions
- Denaturing step: 5 min at 94°C
- 35 cycles of: [94°C 30s, 65°C 30s, 72°C 30s]
- Extension step: 7 min at 72°C
Digestion
- Enzyme: HpyCH4V
- Conditions: digest in Cutsmart Buffer for 2 h at 37°C,
Expected Products
- Separate in a 2% agarose gel:
- WAPO-A1b: one band of 200-bp
- WAPO-A1a, WAPO-A1c, WAPO-A1d: one band of 180-bp
References
1. Identification and validation of QTL for grain yield and plant water status under contrasting water treatments in fall-sown spring wheats. Zhang JL, Gizaw SA, Bossolini E, Hegarty J, Howell T, Carter AH, Akhunov E, Dubcovsky J. In: Theoretical and Applied Genetics, 2018, 131:1741–1759. DOI:10.1007/s00122-018-3111-9.
2. Genome-wide association studies for yield-related traits in soft red winter wheat grown in Virginia. Ward BP, Brown-Guedira G, Kolb FL, Van Sanford DA, Tyagi P, Sneller CH, Griffey CA. In: PLoS ONE, 2019, 14:e0208217. DOI:10.1371/journal.pone.0208217.
3. Genetic architecture of male floral traits required for hybrid wheat breeding. Boeven PHG, Longin CFH, Leiser WL, Kollers S, Ebmeyer E, Wurschum T. In: Theoretical and Applied Genetics 129:2343–2357. DOI:10.1007/s00122-016-2771-6.
4. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Wurschum T, Leiser WL, Langer SM, Tucker MR, Longin CFH. In: Theoretical and Applied Genetics, 2018, 131:2071–2084. DOI:10.1007/s00122-018-3133-3.
5. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Voss-Fels KP, Keeble-Gagnère G, Hickey LT, Tibbits J, Hayden M, Pasam RK, Friedt W, Snowdon RJ, Appels R, Wittkop B. In: Theoretical and Applied Genetics, 2019, 132:2707–2719. DOI:10.1007/s00122-019-03383-4.
6. Identification of quantitative trait loci controlling agronomic traits indicates breeding potential of Tibetan semiwild wheat (Triticum aestivum ssp. tibetanum). Luo W, Ma J, Zhou XH, Sun M, Kong XC, Wei YM, Jiang YF, Qi PF, Jiang QT, Liu YX, Peng YY, Chen GY, Zheng YL, Liu CJ, Lan XJ. In: Crop Science, 2016, 56:2410–2420. DOI:10.2135/cropsci2015.11.0700.
7. Mapping QTLs for yield and nitrogen-related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression. Xu YF, Wang RF, Tong YP, Zhao HT, Xie QG, Liu DC, Zhang AM, Li B, Xu HX, An DG. Theoretical and Applied Genetics, 2014, 127:59–72. DOI:10.1007/s00122-013-2201-y.
8. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Zhai HJ, Feng ZY, Li J, Liu XY, Xiao SH, Ni ZF, Sun QX. In: Frontiers in Plant Science, 2016, 7:1617. DOI:10.3389/fpls.2016.01617.
9. Analysis of agronomic and domestication traits in a durum × cultivated emmer wheat population using a high-density single nucleotide polymorphism-based linkage map. Faris JD, Zhang Q, Chao S, Zhang Z, Xu SS. In: Theoretical and Applied Genetics, 2014, 127:2333–2348. DOI:10.1007/s00122-014-2380-1.
10. Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Kuzay S, Xu Y, Zhang J, Katz A, Pearce S, Su Z, Fraser M, Anderson JA, Brown-Guedira G, DeWitt N, Peters Haugrud A, Faris JD, Akhunov E, Bai G, Dubcovsky J. In: Theoretical and Applied Genetics, 2019, 132, 2689–2705. DOI:10.1007/s00122-019-03382-5.
