Contributed by Gabriela Tranquilli (tranquilli.gabriela@inta.gob.ar)
Wheat marketing systems established a primary classification of hexaploid wheat (Triticum aestivum L.) based on endosperm texture, i.e. the hardness or softness of the grain, because this trait determines many of its potential end-uses. Hard textured grains require more grinding energy than soft textured grains to reduce endosperm into flour, and during this milling process a larger number of starch granules become physically damaged. Since damaged starch granules absorb more water than undamaged granules, flours from hard wheats are preferred for yeast-leavened bread baking, while flours from soft wheats are preferred for manufacturing cookies and cakes.
Endosperm texture is primary controlled by the Hardness (Ha) locus on the short arm of chromosome 5D. It is a simply inherited character and although the main locus is referred as hardness, softness is in fact the dominant trait.
The lipid binding proteins puroindolines a (PINA) and b (PINB) have been identified as responsible in determining differences between hard and soft textured wheats. These proteins are coded by the Pina-D1 and Pinb-D1 genes, tightly linked to the Ha locus on chromosome 5DS (reviewed in 1, 2).
These two genes act together in the determination of grain texture. The presence of wild-type puroindoline alleles at both loci (Pina-D1a and Pinb-D1a) determines softness, while either a deletion resulting in the complete lack of PINA protein, or single -nucleotide mutations resulting in a modified amino acid sequence or null expression of the PINB protein were shown to be inseparably linked to hard-textured grains in surveys of American and European wheats (3-6). It has been observed that genotypes having the deletion at Pina-D1 were harder than those having the single nucleotide mutation in Pinb-D1, and that genotypes having deleted both Pina-D1 and Pinb-D1 were even harder (7, 8). On the other hand, the incorporation of additional copies of Pina-Am1 and Pinb-Am1 from T. monococcum in hexaploid wheat resulted in significantly softer grains than those from the soft control (8). Complementation tests using over-expression of the pinB gene in transgenic wheat plants carrying the soft pinB-D1b allele demonstrated that the puroindoline genes are responsible for the soft texture (9).
Small modulations of this trait might also be achieved by manipulating minor QTLs like the one reported by Campbell et al (10) on chromosome 3AS.
PCR markers for Pina and Pinb
There are PCR-based markers for each puroindoline gene of Triticum aestivum. These are PCR specific primers and CAPs markers. DNA can be isolated by using a micro-scale method, but it is convenient to choose one yielding DNA of good quality, otherwise there might be problems with the amplification.
Pina-D1
Gautier et al (11) developed a dominant PCR marker for this gene.
Primers
Pina-D1-F: 5'- CCC TGT AGA GAC AAA GCT AA -3'
Pina-D1-R: 5'- TCA CCA GTA ATA GCC AAT AGT G -3'
Final concentrations of the products used in the PCR reaction:
- 1X Taq buffer (Mg free)
- dNTP: 250 µM each
- MgCl2: 1.5 mM
- primers: 1 ng/µL each
- Taq polymerase: 2.5 U
- DNA input: 50-100 ng/reaction
Final volume: 25 µl
PCR conditions (times for each step might vary according to the Thermocycler model):
- Denaturing step: 94°C, 3 min
- Amplification step (37 cycles)
- 94°C 90 sec
- 55°C 90 sec
- 72°C 2 min]
- Extension step: 72°C 10 min
Expected products
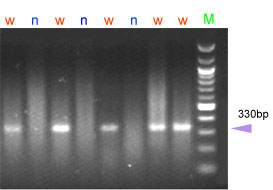
PCR products are visualized in 2% agarose gels, stained with ethidium bromide. Amplification is observed only for the wild-type allele, yielding a 330bp fragment.
| w = wild-type allele (Pina-D1a) n = Null allele (Pina-D1b), associated with hard texture |
 |
Pinb-D1 alleles:
Pinb-D1b: To trace the specific allele for the glycine-to-serine mutation (Pinb-D1b) there are two PCR-based markers. The first one, developed by Giroux and Morris (3,4) uses specific primers for the mutation. However, discrimination of these primers is based on a single base pair difference at the 3’ end of the primers and occasional false positives are observed when PCR conditions are not perfectly optimized. To avoid this problem a codominant PCR-CAPs marker, based on the same point mutation, was developed (12).
After amplification of a 447-bp segment of the Pinb-D1 gene using the primers designed by Gautier et al. (11), the PCR product is digested with the restriction enzyme Bsr BI. This enzyme recognizes the sequence with the glycine-to-serine mutation but not the sequence without the mutation present in all soft varieties. After digestion, fragments of 320-bp are expected for genotypes lacking the glycine to-serine mutation, while fragments of 200-bp are expected for those genotypes carrying the allele for hard texture.
Primers:
PinB-D1-F: 5'- ATG AAG ACC TTA TTC CTC CTA -3'
PinB-D1-R: 5'- TCA CCA GTA ATA GCC ACT AGG GAA -3'
Final concentrations of the products used in the PCR reaction:
- 1X Taq buffer (Mg free)
- dNTP: 250 uM each
- MgCl2: 1.5 mM
- primers: 1 ng/ µL each
- Taq polymerase: 2.5 U
- DNA input: 50-100 ng/reaction
Final volume: 50 µl
PCR conditions (times for each step might vary according to the thermocycler model):
- Denaturing step: 94°C, 3 min
- Amplification step (37 cycles)
- 94°C 90 sec
- 55°C 90 sec
- 72°C 2 min
- Extension step: 72°C 10 min
Restriction:
To perform the restriction, 5U of Bsr BI is added to 25µL of the PCR product and incubated at 37 °C for 4hs. No extra buffers are added.
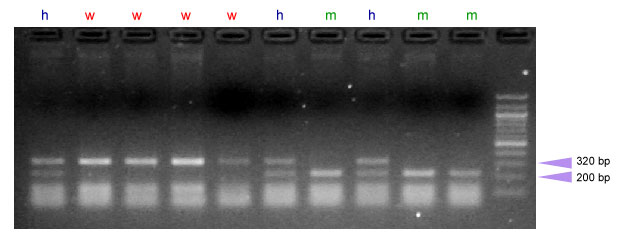 |
| w = wild-type sequence, m = glycine to serine mutation (allele Pinb-D1b), hard texture, h= heterozygous |
Pinb-D1c: A CAPs marker is available to introgress this allele (5). Amplification is performed as indicated for the Pinb-D1b allele. To detect the Pro-60 mutation, the PCR product is digested with 2 U of the restriction enzyme Pvu II.
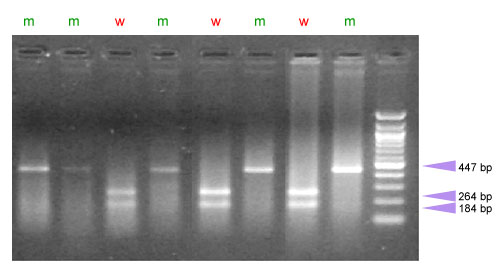 |
| w = wild-type sequence, m = Leucine to proline mutation (allele Pinb-D1c), hard texture, h= heterozygous |
References
1. Puroindolines: the molecular basis of wheat grain hardness. Morris CF In: Plant Molecular Biology, 2002, 48: 633-647. DOI:10.1023/A:1014837431178
2. Endosperm texture in wheat. Turnbull KM, Rahman S. In: Journal of Cereal Science, 2002, 36: 327-337. DOI:10.1006/jcrs.2002.0468
3. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Giroux MJ, Morris CF. In: Theoretical and Applied Genetics, 1997, 95: 857:864. DOI:10.1007/s001220050636
4. Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Giroux MJ, Morris CF. In: Proceedings of the National Academy of Sciences, 1998, 95: 6262-6266. DOI:10.1073/pnas.95.11.6262
5. A leucine to proline mutation in puroindoline b is frequently present in hard wheats from Northern Europe. Lillemo M, Morris CF. In: Theoretical and Applied Genetics, 2000, 100:1100-1107. DOI:10.1007/s001220051392
6. Prevalence of puroindolines grain hardness genotypes among historically significant North American spring and winter wheats. Morris CF, Lillemo M, Simeone MC, Giroux MJ, Babb SL, Kidwell KK. In: Crop Science, 2001, 41: 218-228. DOI:10.2135/cropsci2001.411218x
7. Association of puroindoline sequence type and grain hardness in hard red spring wheat Giroux MJ, Talbert L, Habernicht DK, Lanning S, Hemphill A, Martin JM. In: Crop Science, 2000, 40: 370-374. DOI:10.2135/cropsci2000.402370x
8. Substitutions and deletions of genes related to grain hardness in wheat and their effect on grain texture. Tranquilli G, Heaton J, Chicaiza O, Dubcovsky J. In: Crop Science, 2002, 42: 1812-1817. DOI:10.2135/cropsci2002.1812
9. Expression of wild-type pinB sequence in transgenic wheat complements a hard phenotype. Beecher B, Bettge A, Smidansky E, Giroux, MJ. In: Theoretical and Applied Genetics, 2002, 105: 870-877. DOI:10.1007/s00122-002-1034-x
10. Quantitative Trait Loci Associated with Kernel Traits in a Soft x Hard Wheat Cross. Campbell KG, Bergman CJ, Gualberto DG, Anderson JA, Giroux MJ, Hareland G, Fulcher RG, Sorrells ME, Finney PL..In: Crop Science, 1999, 39:1184–1195. DOI:10.2135/cropsci1999.0011183X003900040039x
11. Triticum aestivum puroindolines, two cystine-rich seed proteins: cDNA sequence analysis and developmentar gene expression Gautier MF, Aleman ME, Guirao A, Marion D, Joudrier P. In: Plant Molecular Biology, 1994, 25: 43-57. DOI:10.1007/BF00024197
12. Genetic and physical characterization of grain texture-related loci in diploid wheat. Tranquilli G, Lijavetzky D, Muzzi G, Dubcovsky J. In: Molecular and General Genetics, 1999, 262: 846 - 850. DOI:10.1007/s004380051149
