One of the main components of wheat flour is starch, its relative content and chemical composition affect the quality of the products obtained from wheat.
Starch is a glucose polymer composed of two different types of structures: amylose and amylopectin. Amylose is a linear polymer with D-glucosyl units linked by α 1-4 bonds. Amylopectin has a branched structure: it also has α 1-4 bonds but every 20-25 glucosyl there are ramifications due to α 1-6 bonds.
The enzyme responsible for amylose biosynthesis in wheat is the granule-bound starch synthase (GBSS) or waxy protein. The synthesis of amylopectin is more complex and different enzymes are necessary. In wheat there are three genes that code for GBSS : Wx-A1, Wx-D1 and Wx-B1, with chromosomal localizations: 7AS, 7DS and 4AL, respectively.
Some mutants of wheat lack one or more GBSS proteins, they are called partial waxy mutants. In wheat lines with mutations in the three genes coding for GBSS, starch is only composed by amylopectin. When one or two genes are mutated the relative amount of amylopectin in starch increases compared to amylose. The largest effects on amylose content and quality are observed in Wx-B1 mutants, followed by Wx-D1 and Wx-A1 mutants (9).
Uses of partial waxy and waxy flours
Consumers of Udon, one of the noodles produced from wheat, prefer a noodle with a firm surface and soft inside. These characteristics are associated with high swelling volumes and high peak pasting viscosities, which are observed in wheats with reduced amylose content and the presence of null alleles for GBSS (3). The null mutants for theWx-B1 locus are usually preferred for Japanese Udon noodles. A complete waxy mutant could not be used for noodle production, but could be used for other industrial applications.
PCR markers for waxy genes
There are a number of markers available for the different waxy genes and their alleles. Here is the list of markers you can find below:
- Dominant PCR markers for the null alleles of the waxy genes. This marker detects null alleles having deletions in the region spanning exons 4 to 6.
- Codominant marker for the Wx-A1b alleles. This CAPS marker differentiates the normal allele from the null allele present in lines Komugi Nourin, Fugimi Komugi, and the triple null THF7.
- Allelic variation of the Wx-A1 locus. This microsatellite marker is useful to characterize the different functional alleles for Wx-A1 and also works as a recessive marker for the null Wx-D1 allele.
- Codominant markers for the normal an null alleles of the Wx-D1 locus. This marker can be used for breeding partially waxy lines carrying the Wx-D1b allele derived from Bai-Huo.
Dominant PCR markers for the null alleles of the waxy genes
McLauchlan et al. published a paper reviewing and validating a number of PCR markers for the different alleles of the waxy genes. Primer sets #3 and #4 amplify all the three loci and can be used to detect null alleles. They amplify fragments located on exons 4 and 6. Primer pair #4 was used in a waxy wheat breeding program (4). For the breeding program at UC Davis we tested primer pairs #3 and #4. We obtained better and more reproducible results with pair #4. For scoring this marker the authors use radioactive primers and the products are analyzed with a capillary sequencer. As a cheaper alternative for non high-throughput programs we tried agarose 3% and the bands can be clearly discriminated.
This PCR protocol was originally described in Ref. 4.
Primers:
Left primer: 5'-AAG AGC AAC TAC CAG T-3'
Right primer: 5'-TCG TAC CCG TCG ATG AAG TCG A-3'
The MgCl2 concentration is 1.5 mM.
PCR program:
- Denaturing step: 95ºC, 3 min
- Touchdown step:
- 94ºC, 1 min
- 64ºC to 58ºC (1ºC/2cycles), 1 min
- 72ºC, 1 min
- Amplification step (25 cycles):
- 94ºC, 1 min
- 58ºC, 1 min
- 72ºC, 30 sec
- Extension step: 72ºC, 5 min
- Hold at 4ºC
Some labs reported problems to reproduce this protocol. These modifications yield stronger bands:
- The MgCl2 concentration was increased to 3 mM.
- The amplification step has 35 cycles at 58ºC, instead of 25 cycles.
- Final primer concentration: 0.2 µM each.
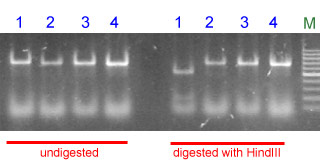
The picture below shows a typical result for the modified protocol. Gamenya and Haelberg are WxB1b lines.
 |
Codominant CAPS markers for the null allele Wx-A1b
Contributed by Marcelo Helguera (helguera.marcelo@inta.gob.ar)
This codominant marker differentiates the normal Wx-A1a allele from the null Wx-A1b allele present in lines Komugi Nourin, Fugimi Komugi and the triple mutant THF7. After the PCR reaction a slight difference in product sizes can be observed, but the treatment with the restriction enzyme improves the visualization in an agarose gel.
Primers:
Wx-A1b-F-MH 5'-CCC CAA AGC AAA GCA GGA AAC-3'
Wx-A1b-R-MH 5'-CGG CGT CGG GTC CAT AGA TC-3'
PCR reaction:
- MgCl2: 1.5 mM
- dNTP: 200 µM each
- DNA: 120 ng/reaction
- Taq Polymerase (Promega): 1U/reaction (0.2µl/reaction)
final volume: 25 µl
PCR program:
- Denaturing step: 94ºC, 3 min
- Amplification step (40 cycles):
- 94ºC, 45 sec
- 55ºC, 30 sec
- 72ºC, 60 sec
- Extension step: 72ºC, 10 min
- Hold at 4ºC
Digestion with Hind III (Promega):
Add 5U of the enzyme to 10 µl of the PCR reaction and incubate at 37°C for 1 h.
|
|
|
Allelic variation of the Wx-A1 locus.
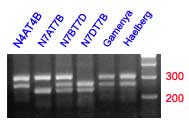
Sharoflou and Sharp (6) found a microsatellite sequence near the 3' end of a waxy cDNA sequence and designed PCR primers flanking this region (Accession X57233). The PCR amplification of Chinese Spring DNA produced two bands, one of 204 bp and the other of 265 bp. Using aneuploid stocks the 204 bp product was assigned to chromosome 7D (locus XSun1-7D) and the 265 bp product to chromosome 7A (locus XSun1-7A). This region is particularly useful for analyzing the allelic variation of the Wx-A1 locus (4,6).
Both loci were analyzed in 135 varieties. The XSun1-7A locus is highly polymorphic, with 8 alleles detected. Two varieties that were null for the Wx-A1 gene gave unique fragments of 233 bp. The XSun1-7D locus was not polymorphic in the lines analyzed, but the expected fragment of 204 bp was lacking in varieties null for the Wx-D1 gene.
Primers:
Sun1F: 5'-CGC TCC CTG AAG AGA GAA AGA A-3'
Sun1R: 5'-ATA GGC ACA ACC CCT AAC -3'
PCR conditions:
- Denaturing step: 95°C, 3 min
- 5 cycles of: [94°C 1 min, 58°C 1 min, 72°C 1 min]
- 25 cycles of: [94°C 1 min, 58°C 50 sec, 72°C 30 sec]
- Extension step: 72°C 4 min
Final volume: 25 μl
Final concentrations of the products used in the PCR reaction:
- dNTPs: 200 μM each
- MgCl2: 1.5 mM
- primers: 0.5 μM each
- Taq DNA polymerase (Advanced Biotechnologies, Epson, UK): 1 unit
- 1X PCR buffer (Advanced Biotechnologies, Epson, UK)
- Genomic DNA: 75 ng
Expected products
Amplification products were separated in 3% agarose and ethidium bromide staining or 6% polyacrylamide with silver staining.
Eight different alleles were detected at the Xsun-7A locus in 135 cultivars. Their sizes were: 219, 233, 260, 271, 275, 285 and 289 bp. Locus Xsun-7D is not polymorphic. However, it is a recessive marker for null alleles of the Wx-D1 gene, since no amplification was observed for the 3 Wx-D1 null varieties analyzed. For the rest of the varieties the product obtained had a size of 204 bp.
Codominant markers for the normal and null allele of the Wx-D1 locus
Baihuomai is a Chinese landrace with a null allele, Wx-D1b, at the Wx-D1 locus. This line could be the same as Bai-Huo, which we are using for some of our breeding programs. Vrinte et. al (7) and Shariflou et al. (10) developed markers for this allele. Both methods are described below.
PCR protocol for the marker developed by Shariflou et. al (10)
Primers:
Wx-D1-2-F 5'-ACA GGA TCT CTC CTG GAA G-3'
Wx-D1-2-R 5'-GCA AGG AAA ATA GTG AAG C-3'
PCR reaction:
- primers: 0.5 µM each
- MgCl2: 1.5 mM
- dNTP: 200 µM each
- DNA: 100 ng/reaction
- Taq Polymerase (Promega): 1U/reaction (0.2µl/reaction)
final volume: 25 µl
PCR program:- Denaturing step: 95ºC, 3 min
- Touchdown step (8 cycles):
- 94ºC, 1 min
- 63ºC to 55ºC 1ºC/cycle, 1 min
- 72ºC, 1 min
- Amplification step (33 cycles):
- 94ºC, 1 min
- 55ºC, 1 min
- 72ºC, 1 min
- Extension step: 72ºC, 5 min
- Hold at 4ºC
The null allele of cultivar Bai-Huio yields a band of 279 bp, while the amplification product from the normal allele is larger (see figure below).
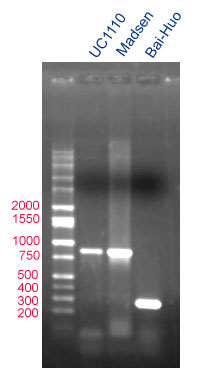 |
PCR protocol for the marker developed by Vrinte et. al (7)
Primers:
Wx-D1-1-F 5'-GAG ATG GTC AAG AAC TGC AT-3' Wx-D1-1-R 5'-TAG TGC GTC CAG ACT CAC AG-3'
PCR reaction:- primers: 0.5 µM each
- MgCl2: 1.5 mM
- dNTP: 200 µM each
- DNA: 100 ng/reaction
- Taq Polymerase (Promega): 1U/reaction (0.2µl/reaction)
final volume: 25 µl
PCR program:- Denaturing step: 95ºC, 3 min
- Touchdown step (8 cycles):
- 94ºC, 1 min
- 65ºC to 57ºC 1ºC/cycle, 1 min
- 72ºC, 1 min
- Amplification step (25 cycles):
- 94ºC, 1 min
- 55ºC, 1 min
- 72ºC, 1 min
- Extension step: 72ºC, 5 min
- Hold at 4ºC
References
1. A PCR-based marker for selection of starch and potential noodle quality in wheat. Briney, A; Wilson, R.; Potter, R. H.; Barclay, I.; Crosbie, G.; Appels, R.; Jones, M.G. J. In: Molecular Breeding, 1998, 4(5):427-433. DOI:10.1023/A:1009664917998
2. Application of a high-throughput antibody-based assay for identification of the granule-bound starch synthase Wx-B1b allele in Australian wheat lines. Gale, K. R.; Panozzo, J. F.; Eagles, H. A.; Blundell, M.; Olsen, H.; Appels, R. In: Australian Journal of Agricultural Research, 2001, 52(11-12):1417-1423. DOI:10.1071/AR01037
3. Waxy wheats: Origin, properties, and prospects Graybosch, R.A. In: Trends in Food Science & Technology, 1998, 9(4):135-142. DOI:10.1016/S0924-2244(98)00034-X
4. Development of robust PCR-based DNA markers for each homoeo-allele of granule-bound starch synthase and their application in wheat breeding programs. McLauchlan, A.; Ogbonnaya, F. C.; Hollingsworth, B.; Carter, M.; Gale, K.R.; Henry, R.J.; Holton, T.A.; Morell, M. K.; Rampling, L.R.; Sharp, P. J.; Shariflou, M. R.; Jones, M.G.K.; Appels, R. In: Australian Journal of Agricultural Research, 2001, 52(11-12):1409-1416. DOI:10.1071/AR01036
5. Registration of D-null "Bai Huo" waxy wheat germplasm. . Morris, C.F.; Konzak, C.F. In: Crop Science, 2000, 40(1):304-305. DOI:10.2135/cropsci2000.0011rgp
6. A polymorphic microsatellite in the 3' end of 'waxy' genes of wheat, Triticum aestivum. Shariflou, M. R.; Sharp, P. J. In: Plant Breeding, 1999, 118(3):275-277. DOI:10.1046/j.1439-0523.1999.118003275.x
7. Molecular characterization of waxy mutations in wheat. Vrinten, P.; Nakamura, T.; Yamamori M. In: Molecular and General Genetics, 1999, 261(3):463-471. DOI:10.1007/s004380050989
8. The genes encoding granule-bound starch synthases at the waxy loci of the A, B, and D progenitors of common wheat. Yan L.; Bhave M.; Fairclough R.; Konik C.; Rahman S.; Appels R. In: Genome, 2000, 43(2):264-272. DOI:10.1139/g99-117
9. Differential effects of Wx-A1,-B1 and-D1 protein deficiencies on apparent amylose content and starch pasting properties in common wheat. Yamamori, M.; Quynh, N.T. In: Theoretical and Applied Genetics, 2000, 100:32-38. DOI:10.1007/s001220050005
10. A PCR-based DNA marker for detection of mutant and normal alleles of the Wx-D1 gene of wheat. Shariflou M.R.; Hassani M.E.; Sharp P.J. In: Plant Breeding, 2001, 120(2):121-124. DOI:10.1046/j.1439-0523.2001.00577.x
11. Production of all eight genotypes of null alleles at 'waxy' loci in bread wheat, Triticum aestivum L. Zhao, X.C.; Sharp, P.J., In: Plant Breeding, 1998, 117:488-490. DOI:10.1111/j.1439-0523.1998.tb01979.x
12. Isolation and characterization of the three Waxy genes encoding the granule-bound starch synthase in hexaploid wheat. Murai J, Taira T, Ohta D. In: Gene, 1999, 234(1):71-79. DOI:10.1016/S0378-1119(99)00178-X
13. Nucleotide sequence of a wheat (Triticum aestivum L.) cDNA clone encoding the waxy protein. Clark JR, Robertson M, Ainsworth CC. In: Plant Molecular Biology, 1991, 16(6):1099-1101. DOI:10.1007/BF00016086