Grain protein content (GPC) is one of the most important quality factors for pasta and bread wheats. Breeding for an increase in GPC is difficult because the genetic variation for this character is small compared to variations caused by the environment, besides there is a negative correlation between GPC and grain yield.
The high grain protein content (Gpc-B1) gene transferred from Triticum turgidum ssp. dicoccoides into durum wheat is a valuable resource for increasing GPC (1, 2). Some results suggest that in different backgrounds this gene has none or minimal effects on grain yield, protein quality, plant height or heading date (2, 4). The Gpc-B1 gene was also transferred to hexaploid backgrounds. One of the hexaploid lines obtained was Glupro, which is the donor parent in several backcrossing programs within the IFAFS project. Different mapping experiments indicated that the location of the QTL for the transferred Gpc-B1 gene in both, pasta and bread wheat, is on chromosome arm 6BS, between the Nor and the centromere (3, 5, 6).
The stripe rust resistance gene Yr36 is closely linked to the Gpc locus, and the same markers used for tagging Gpc-B1 can be used for Yr36.
Markers for Gpc-B1 and Yr36
Xuhw89
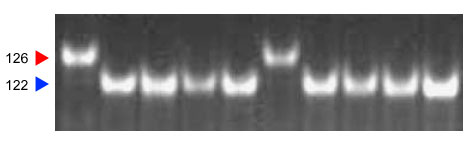
This PCR marker is located very close (0.1-cM) of the Gpc-B1 gene, and can be used in high-throughput systems (8). It reveals a 4-bp deletion linked to the Gpc-B1 gene from Triticum turgidum ssp. dicoccoides, and is absent in most common and durum wheat lines. None of the 117 varieties included in a screening with Xuhw89 showed the 4-bp deletion (8).
Primers to detect the 4-bp deletion:
UHW89-BF 5'- TCT CCA AGA GGG GAG AGA CA -3'
UHW89-R 5'- TTC CTC TAC CCA TGA ATC TAG CA -3'
Final concentrations of the products used in the PCR reaction:
- dNTP: 200 µM each
- primers: 0.2 µM each
- 1X Taq buffer (Mg2+ free)
- MgCl2 1.5 mM
- Taq polymerase: 2.5 U
- DNA input: 50-100 ng/reaction
Final volume: 25 µl
PCR and restriction conditions:
- Denaturing step: 5 min at 94°C
- 37 cycles of: [94°C 30s, 59°C 30s, 72°C 45s]
- Extension step: 5 min at 72°C
Expected products:
Amplification products were separated on a 6% polyacrylamide gel.
Xucw71 and Xucw79
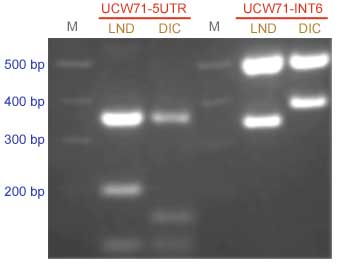
The Gpc-B1 locus was mapped within a 0.3-cM interval defined by flanking loci Xucw71 and and Xucw79 (7). Since the distance between the Gpc-B1 gene and these two markers is small, selection can be done using a single marker, limiting the use of the second markers to the last cycle of selection to confirm the correct transfer of the Gpc-B1 gene. The Xucw71 marker has been validated in a wide range of germplasms
Xucw71 (Ca2+/H+ antiporter-related protein)
Distelfeld et al. (7) developed two PCR-CAPS markers for the Xucw71 locus. These markers are based on SNPs (single nucleotide polymorphisms) on a wheat locus with sequence homology to a rice Ca2+/H+ antiporter-related protein. The first SNP is located on the 5' UTR and the second on intron 6 of the gene. Both SNPs are detected with the same restriction enzyme Bsm I.
Primers for the first SNP in the 5’ UTR (5'UTR SNP):
UCW71_5UTR_F 5'- CTT GCA CCC GTG GAT CAG -3'
UCW71_5UTR_R 5'- CGA TGC AAT AAT TTA TCA CAC GTA -3'
Primers for the second SNP in intron six (INTRON 6 SNP):
UCW71_INT6_F 5'- TGG ACT TTC TAT TTC TCC GTA CC -3
UCW71_INT6_R 5'- TCA ACC CTT TTA AGC AAT TTG AA -3
Final concentrations of the products used in the PCR reaction:
- dNTP: 200 µM each
- primers: 0.16 µM each
- 1X Taq buffer (Mg2+ free)
- Taq polymerase: 2.5 U
- DNA input: 50-100 ng/reaction
Final volume: 25 µl
PCR and restriction conditions:
- Denaturing step: 5 min at 94°C
- Touchdown from 53° to 48° (six cycles).
- Denaturation: 30s at 94°
- Annealing: 30s at {53, 52, 51, 50, 49, 48 °C} in each cycle
- Extension at 72 ° for 40 seconds
- 37 cycles of: [94°C 30s, 48°C 30s, 72°C 45s]
- Extension step: 5 min at 72°C
Digestion with Bsm I : 15 µl of PCR product are digested with 0.5 µl (5 units) of Bsm I (New England Biolabs) plus 1.5 µL of water. Bsm I works directly in PCR buffer. The reaction mixture is incubated at 65°C 3 hours.
Expected products:
Restriction products were separated on a 2% agarose gel.
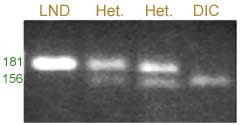
Xucw79 (heterotrimeric G-protein gamma subunit 2 )
Primers
UCW79-dCAPSF 5'- AGA TAA CGA CCG ATG CGA TCT TAG TA -3'
UCW79-dCAPSR 5'- TCC TTT TTC CGA TTT TCT TTG TGT -3'
The degenerate T in the second base from the 3’ end (TCT TAG TA) generates an Acc I restriction site with the SNP present in DIC.
The components and concentrations for the PCR reaction are as for Xucw71
PCR and restriction conditions:
- Denaturing step: 5 min at 94°C
- Touchdown from 55° to 50° (six cycles).
- Denaturation: 30s at 94°
- Annealing: 30s at {55, 54, 53, 52, 51, 50°C} in each cycle
- Extension at 72 ° for 40 seconds
- 37 cycles of: [94°C 30s, 50°C 30s, 72°C 45s]
- Extension step: 5 min at 72°C
Digestion with Acc I : 15 µl of PCR product are digested with 0.5 µl (5 units) of Acc I (New England Biolabs) plus 1.5 µL of water. Acc I works directly in PCR buffer. The reaction mixture is incubated at 37°C 3 hours.
Expected products:
Restriction products were separated on a 2.5% agarose gel.
References
1. Chromosomal location of genes for grain protein content of wild tetraploid wheat. Joppa LR, Cantrell RG. In: Crop Science, 1990, 30(5):1059-1064. DOI:10.2135/cropsci1990.0011183X003000050021x.
2. Grain protein determinants of the Langdon durum-dicoccoides chromosome substitution lines. Deckard EL, Joppa LR, Hammond JJ, Hareland GA. In: Crop Science, 1996, 36(6):1513-1516. DOI:10.2135/cropsci1996.0011183X003600060017x.
3. Mapping gene(s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Joppa LR, Du C, Hart GE, Hareland GA. In: Crop Science, 1997, 37(5):1586-1589. DOI:10.2135/cropsci1997.0011183X003700050030x.
4. Quality characteristics of durum wheat lines deriving high protein from a Triticum dicoccoides (6b) substitution. Kovacs MIP, Howes NK, Clarke JM, Leisle D. In: Journal of Cereal Science, 1998, 27(1):47-51. DOI:10.1006/jcrs.1997.0144.
5. RFLP markers associated with high grain protein from Triticum turgidum L. var. dicoccoides introgressed into hard red spring wheat. Mesfin A, Frohberg RC, Anderson JA. In: Crop Science, 1999, 39(2):508-513. DOI:10.2135/cropsci1999.0011183X003900020035x.
6. Development of PCR-based markers for a high grain protein content gene from Triticum turgidum ssp. dicoccoides transferred to bread wheat. Khan IA, Procunier JD, Humphreys DG, Tranquilli G, Schlatter AR, Marcucci-Poltri S, Frohberg R, Dubcovsky J. In: Crop Science, 2000, 40(2):518-524. DOI:10.2135/cropsci2000.402518x.
7. Microcolinearity between a 2-cM region encompassing the grain protein content locus Gpc-6B1 on wheat chromosome 6B and a 350-kb region on rice chromosome 2. Distelfeld A, Uauy C, Olmos S, Schlatter AR, Dubcovsky J, Fahima T. In: Functional & Integrative Genomics, 2004, 4: 59-66. DOI:10.1007/s10142-003-0097-3.
8. Physical map of the wheat high-grain protein content gene Gpc-B1 and development of a high-throughput molecular marker. Distelfeld A, Uauy C, Fahima T, Dubcovsky J. In: New Phytologist, 2006, 169:753-763. DOI:10.1111/j.1469-8137.2005.01627.x.
